3OH4
 
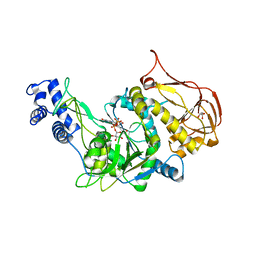 | | Protein structure of USP from L. major bound to URIDINE-5'-DIPHOSPHATE Glucose | | Descriptor: | GLYCEROL, UDP-sugar pyrophosphorylase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F.H, Ficner, R. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
6WP2
 
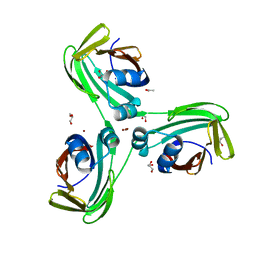 | |
6WZ4
 
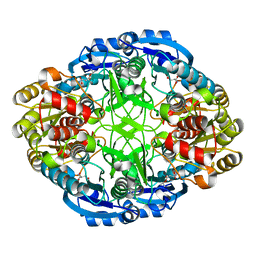 | | Complex of mutant (K173M) of Pseudomonas 7A Glutaminase-Asparaginase with L-Asp at pH 6. Covalent acyl-enzyme intermediate | | Descriptor: | ASPARTIC ACID, Glutaminase-asparaginase | | Authors: | Strzelczyk, P, Zhang, D, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2020-05-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Generalized enzymatic mechanism of catalysis by tetrameric L-asparaginases from mesophilic bacteria.
Sci Rep, 10, 2020
|
|
5MJG
 
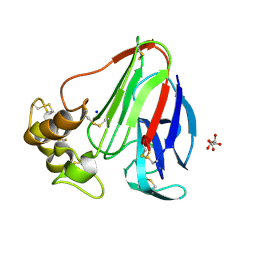 | | Single-shot pink beam serial crystallography: Thaumatin | | Descriptor: | S,R MESO-TARTARIC ACID, SODIUM ION, Thaumatin-1 | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V. | | Deposit date: | 2016-12-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single-shot pink beam serial crystallography: Thaumatin
To Be Published
|
|
7UPZ
 
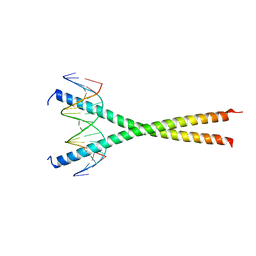 | | Structural basis for cell type specific DNA binding of C/EBPbeta: the case of cell cycle inhibitor p15INK4b promoter | | Descriptor: | CCAAT/enhancer-binding protein beta, DNA (5'-D(*AP*TP*TP*CP*TP*TP*AP*AP*GP*AP*AP*AP*GP*AP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*TP*CP*TP*TP*TP*CP*TP*TP*AP*AP*GP*AP*A)-3') | | Authors: | Lountos, G.T, Cherry, S, Tropea, J.E, Wlodawer, A, Miller, M. | | Deposit date: | 2022-04-18 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structural basis for cell type specific DNA binding of C/EBP beta : The case of cell cycle inhibitor p15INK4b promoter.
J.Struct.Biol., 214, 2022
|
|
3OH1
 
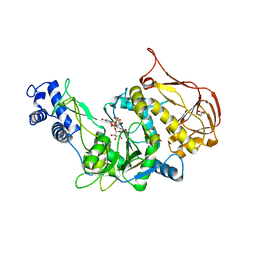 | | Protein structure of USP from L. major bound to URIDINE-5'-DIPHOSPHATE-Galacturonic acid | | Descriptor: | (2S,3R,4S,5R,6R)-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4,5-trihydroxy-oxane-2-carboxylic acid, GLYCEROL, UDP-sugar pyrophosphorylase | | Authors: | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F, Ficner, R. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
6WR4
 
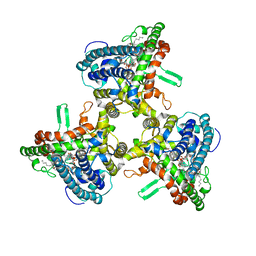 | | Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery | | Descriptor: | Autophagy-related protein 9A, Lauryl Maltose Neopentyl Glycol | | Authors: | Guardia, C.M, Tan, X, Lian, T, Rana, M.S, Zhou, W, Christenson, E.T, Lowry, A.J, Faraldo-Gomez, J.D, Bonifacino, J.S, Jiang, J, Banerjee, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of Human ATG9A, the Only Transmembrane Protein of the Core Autophagy Machinery.
Cell Rep, 31, 2020
|
|
3OH0
 
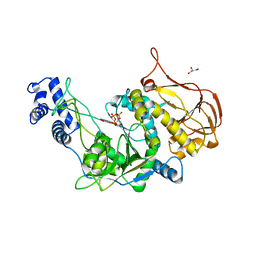 | | Protein structure of USP from L. major bound to URIDINE-5'-TRIPHOSPHATE | | Descriptor: | GLYCEROL, UDP-sugar pyrophosphorylase, URIDINE 5'-TRIPHOSPHATE | | Authors: | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F, Ficner, R. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
6X3X
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus diazepam | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-CHLORO-1-METHYL-5-PHENYL-1,3-DIHYDRO-2H-1,4-BENZODIAZEPIN-2-ONE, ... | | Authors: | Kim, J.J, Gharpure, A, Teng, J, Zhuang, Y, Howard, R.J, Zhu, S, Noviello, C.M, Walsh, R.M, Lindahl, E, Hibbs, R.E. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Shared structural mechanisms of general anaesthetics and benzodiazepines.
Nature, 585, 2020
|
|
6WT4
 
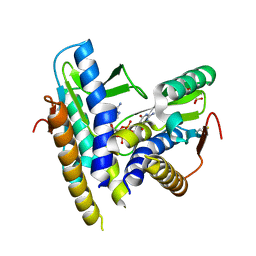 | | Structure of a bacterial STING receptor from Flavobacteriaceae sp. in complex with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Bacterial STING, SULFATE ION | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
6WT7
 
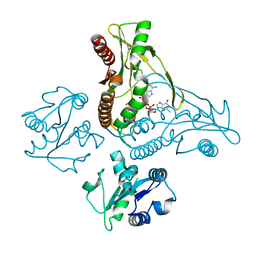 | | Structure of a metazoan TIR-STING receptor from C. gigas in complex with 2',3'-cGAMP | | Descriptor: | Metazoan TIR-STING fusion, cGAMP | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
7UXD
 
 | |
6WYZ
 
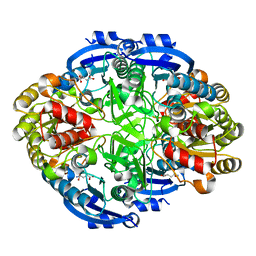 | | Crystal structure of Pseudomonas 7A Glutaminase-Asparaginase (mutant K173M) in complex with D-Glu at pH 5.5 | | Descriptor: | D-GLUTAMIC ACID, Glutaminase-asparaginase | | Authors: | Strzelczyk, P, Zhang, D, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2020-05-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Generalized enzymatic mechanism of catalysis by tetrameric L-asparaginases from mesophilic bacteria.
Sci Rep, 10, 2020
|
|
2PHT
 
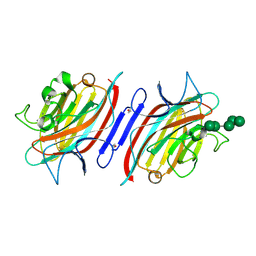 | | Pterocarpus angolensis lectin (P L) in complex with Man-7D3 | | Descriptor: | CALCIUM ION, Lectin, MANGANESE (II) ION, ... | | Authors: | Garcia-Pino, A, Buts, L, Wyns, L, Imberty, A, Loris, R. | | Deposit date: | 2007-04-12 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How a plant lectin recognizes high mannose oligosaccharides
Plant Physiol., 144, 2007
|
|
2PQQ
 
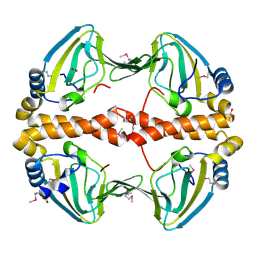 | | Structural Genomics, the crystal structure of the N-terminal domain of a transcriptional regulator from Streptomyces coelicolor A3(2) | | Descriptor: | FORMIC ACID, Putative transcriptional regulator | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-02 | | Release date: | 2007-06-05 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the N-terminal domain of a transcriptional regulator from Streptomyces coelicolor A3(2)
To be Published
|
|
6VYB
 
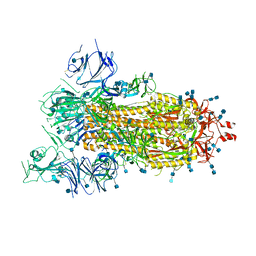 | | SARS-CoV-2 spike ectodomain structure (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Walls, A.C, Park, Y.J, Tortorici, M.A, Wall, A, Seattle Structural Genomics Center for Infectious Disease (SSGCID), McGuire, A.T, Veesler, D. | | Deposit date: | 2020-02-25 | | Release date: | 2020-03-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein.
Cell, 181, 2020
|
|
6RV4
 
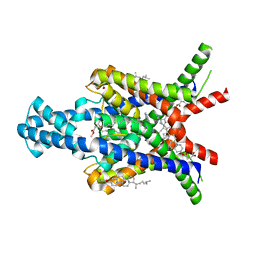 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 2341237 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, POTASSIUM ION, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
6VOC
 
 | | icosahedral symmetry reconstruction of brome mosaic virus (RNA 3+4) | | Descriptor: | Capsid protein | | Authors: | Beren, C, Cui, Y.X, Chakravarty, A, Yang, X, Rao, A.L.N, Knobler, C.M, Zhou, Z.H, Gelbart, W.M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Genome organization and interaction with capsid protein in a multipartite RNA virus.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5NET
 
 | |
7UUR
 
 | | The 1.67 Angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, HYDROXIDE ION, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-04 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (1.67 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
6W0B
 
 | | Open-gate KcsA soaked in 2 mM BaCl2 | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.604 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
3OS6
 
 | | Crystal structure of putative 2,3-dihydroxybenzoate-specific isochorismate synthase, DhbC from Bacillus anthracis. | | Descriptor: | GLYCEROL, Isochorismate synthase DhbC, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Domagalski, M.J, Chruszcz, M, Skarina, T, Onopriyenko, O, Cymborowski, M, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-08 | | Release date: | 2010-10-20 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of isochorismate synthase DhbC from Bacillus anthracis.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6VUZ
 
 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT353 | | Descriptor: | 2-{[3-(piperidin-1-yl)propyl]sulfanyl}-6,7-dihydro-5H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-amine, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Punetha, A, Hou, C, Ngo, H.X, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Guided Optimization of Inhibitors of Acetyltransferase Eis fromMycobacterium tuberculosis.
Acs Chem.Biol., 15, 2020
|
|
2PPL
 
 | | Human Pancreatic lipase-related protein 1 | | Descriptor: | CALCIUM ION, Pancreatic lipase-related protein 1, SODIUM ION | | Authors: | Walker, J.R, Davis, T, Seitova, A, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-30 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Human Pancreatic Lipase-related Protein 1.
To be Published
|
|
7U1Y
 
 | | Structure of SPAC806.04c protein from fission yeast bound to AlF4 and Co2+ | | Descriptor: | COBALT (II) ION, Damage-control phosphatase SPAC806.04c, POTASSIUM ION, ... | | Authors: | Jacewicz, A, Sanchez, A.M, Shuman, S. | | Deposit date: | 2022-02-22 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fission yeast Duf89 and Duf8901 are cobalt/nickel-dependent phosphatase-pyrophosphatases that act via a covalent aspartyl-phosphate intermediate.
J.Biol.Chem., 298, 2022
|
|
