6C5J
 
 | | S25-23 Fab in complex with Chlamydiaceae LPS (Crystal form 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-beta-D-glucopyranose-(1-6)-2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, IgG1 Fab Heavy Chain, ... | | Authors: | Haji-Ghassemi, O, Evans, S.V. | | Deposit date: | 2018-01-16 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Subtle Changes in the Combining Site of the Chlamydiaceae-Specific mAb S25-23 Increase the Antibody-Carbohydrate Binding Affinity by an Order of Magnitude.
Biochemistry, 58, 2019
|
|
2BE9
 
 | | Crystal structure of the CTP-liganded (T-State) aspartate transcarbamoylase from the extremely thermophilic archaeon Sulfolobus acidocaldarius | | Descriptor: | Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | De Vos, D, Savvides, S.N, Van Beeumen, J.J. | | Deposit date: | 2005-10-23 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Sulfolobus acidocaldarius aspartate carbamoyltransferase in complex with its allosteric activator CTP.
Biochem.Biophys.Res.Commun., 372, 2008
|
|
6C5H
 
 | | S25-5 Fab in complex with Chlamydiaceae-specific LPS antigen | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-beta-D-glucopyranose-(1-6)-2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Evans, S.V, Haji-Ghassemi, O. | | Deposit date: | 2018-01-16 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Subtle Changes in the Combining Site of the Chlamydiaceae-Specific mAb S25-23 Increase the Antibody-Carbohydrate Binding Affinity by an Order of Magnitude.
Biochemistry, 58, 2019
|
|
6C5K
 
 | | S25-23 Fab in complex with Chlamydiaceae LPS (Crystal form 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-beta-D-glucopyranose-(1-6)-2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, IgG1 Fab Heavy Chain, ... | | Authors: | Haji-Ghassemi, O, Evans, S.V. | | Deposit date: | 2018-01-16 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Subtle Changes in the Combining Site of the Chlamydiaceae-Specific mAb S25-23 Increase the Antibody-Carbohydrate Binding Affinity by an Order of Magnitude.
Biochemistry, 58, 2019
|
|
1G9K
 
 | |
6E57
 
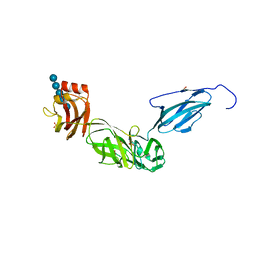 | | Bacteroides ovatus mixed-linkage glucan utilization locus (MLGUL) SGBP-B in complex with mixed-linkage heptasaccharide | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Koropatkin, N.M, Schnizlein, M, Bahr, C.M.E. | | Deposit date: | 2018-07-19 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Surface glycan-binding proteins are essential for cereal beta-glucan utilization by the human gut symbiont Bacteroides ovatus.
Cell.Mol.Life Sci., 76, 2019
|
|
6GDL
 
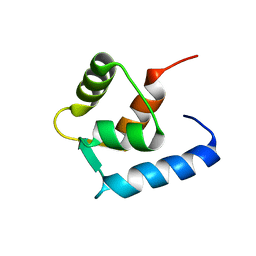 | |
6GDK
 
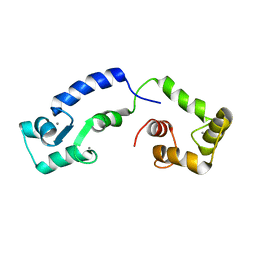 | | Calcium bound form of human calmodulin mutant F141L | | Descriptor: | CALCIUM ION, Calmodulin-1 | | Authors: | Grachov, O, Holt, C, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2018-04-23 | | Release date: | 2018-10-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Arrhythmia mutations in calmodulin cause conformational changes that affect interactions with the cardiac voltage-gated calcium channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4ESR
 
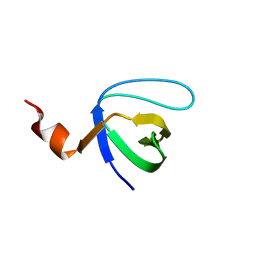 | | Molecular and Structural Characterization of the SH3 Domain of AHI-1 in Regulation of Cellular Resistance of BCR-ABL+ Chronic Myeloid Leukemia Cells to Tyrosine Kinase Inhibitors | | Descriptor: | DI(HYDROXYETHYL)ETHER, Jouberin | | Authors: | Van Petegem, X.F, Liu, P.X, Lobo, P, Jiang, X. | | Deposit date: | 2012-04-23 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Molecular and structural characterization of the SH3 domain of AHI-1 in regulation of cellular resistance of BCR-ABL(+) chronic myeloid leukemia cells to tyrosine kinase inhibitors.
Proteomics, 12, 2012
|
|
6DMF
 
 | |
2MC2
 
 | |
7E0Z
 
 | | Crystal structure of PKAc-PLN complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLN, ... | | Authors: | Qin, J, Yuchi, Z. | | Deposit date: | 2021-01-28 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structures of PKA-phospholamban complexes reveal a mechanism of familial dilated cardiomyopathy.
Elife, 11, 2022
|
|
7E12
 
 | | Crystal structure of PKAc-A11E complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, THR-ARG-SER-GLU-ILE-ARG-ARG-ALA-SER-THR-ILE-GLU, ... | | Authors: | Qin, J, Lin, L, Yuchi, Z. | | Deposit date: | 2021-01-28 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structures of PKA-phospholamban complexes reveal a mechanism of familial dilated cardiomyopathy.
Elife, 11, 2022
|
|
7E11
 
 | | Crystal structure of PKAc-PLN R9C complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLN, ... | | Authors: | Qin, J, Lin, L, Yuchi, Z. | | Deposit date: | 2021-01-28 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Structures of PKA-phospholamban complexes reveal a mechanism of familial dilated cardiomyopathy.
Elife, 11, 2022
|
|
