5FAA
 
 | | Crystal structure of C-terminal domain of shaft pilin spaA from Lactobacillus rhamnosus GG, - I422 space group | | Descriptor: | 1,2-ETHANEDIOL, Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-11 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
5FGS
 
 | | Crystal structure of C-terminal domain of shaft pilin spaA from Lactobacillus rhamnosus GG - P21212 space group | | Descriptor: | Cell surface protein SpaA, ZINC ION | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-21 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
5FGR
 
 | | Crystal structure of C-terminal domain of shaft pilin spaA from Lactobacillus rhamnosus GG - P21212 space group with Yb Heavy atom | | Descriptor: | Cell surface protein SpaA, YTTERBIUM (III) ION | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-21 | | Release date: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
5FIE
 
 | | Crystal structure of N-terminal domain of shaft pilin spaA from Lactobacillus rhamnosus GG | | Descriptor: | Cell surface protein SpaA, SODIUM ION | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
8C46
 
 | |
3EG2
 
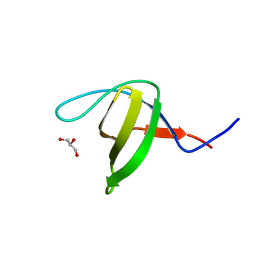 | | Crystal structure of the N114Q mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-10 | | Release date: | 2009-09-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
3EG0
 
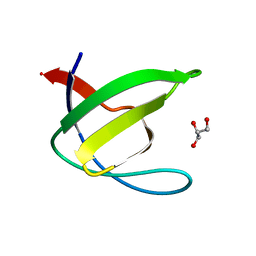 | | Crystal structure of the N114T mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-10 | | Release date: | 2009-09-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
6MAU
 
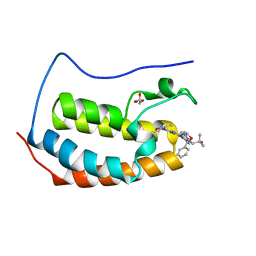 | | Crystal structure of human BRD4(1) in complex with CN210 (compound 19) | | Descriptor: | 1-(4-{6-(3,5-dimethyl-1,2-oxazol-4-yl)-4-[(3S)-3-phenylmorpholin-4-yl]quinazolin-2-yl}-1H-pyrazol-1-yl)-2-methylpropan-2-ol, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Nadupalli, A, Fontano, E, Connors, C.R, Chan, S.G, Olland, A.M, Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2018-08-28 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Lead optimization and efficacy evaluation of quinazoline-based BET family inhibitors for potential treatment of cancer and inflammatory diseases.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
3EG1
 
 | |
3EGU
 
 | | Crystal structure of the N114A mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1, SULFATE ION | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-11 | | Release date: | 2009-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
3EG3
 
 | | Crystal structure of the N114A mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-10 | | Release date: | 2009-09-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
7JY0
 
 | | Structure of HbA with compound 9 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 2-amino-3-{(1S)-1-[5-fluoro-2-(1H-pyrazol-1-yl)phenyl]ethoxy}quinoline-6-carboxamide, CARBON MONOXIDE, ... | | Authors: | Jasti, J. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | PF-07059013: A Noncovalent Modulator of Hemoglobin for Treatment of Sickle Cell Disease.
J.Med.Chem., 64, 2021
|
|
7JXZ
 
 | | Structure of HbA with compound (S)-4 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 3-{(1S)-1-[5-fluoro-2-(1H-pyrazol-1-yl)phenyl]ethoxy}-5-(3-methyl-1H-pyrazol-4-yl)pyridin-2-amine, CARBON MONOXIDE, ... | | Authors: | Jasti, J. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | PF-07059013: A Noncovalent Modulator of Hemoglobin for Treatment of Sickle Cell Disease.
J.Med.Chem., 64, 2021
|
|
7JY3
 
 | | Structure of HbA with compound 23 (PF-07059013) | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 6-{(1S)-1-[(2-amino-6-fluoroquinolin-3-yl)oxy]ethyl}-5-(1H-pyrazol-1-yl)pyridin-2(1H)-one, Hemoglobin subunit alpha, ... | | Authors: | Jasti, J. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | PF-07059013: A Noncovalent Modulator of Hemoglobin for Treatment of Sickle Cell Disease.
J.Med.Chem., 64, 2021
|
|
7JY1
 
 | | Structure of HbA with compound 19 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Jasti, J. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | PF-07059013: A Noncovalent Modulator of Hemoglobin for Treatment of Sickle Cell Disease.
J.Med.Chem., 64, 2021
|
|
8G9Z
 
 | | High-resolution crystal structure of the human selenomethionine-derived SepSecS-tRNASec complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, O-phosphoseryl-tRNA(Sec) selenium transferase, ... | | Authors: | Puppala, A, Simonovic, M, Castillo Suchkou, J. | | Deposit date: | 2023-02-22 | | Release date: | 2023-04-05 | | Last modified: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for the tRNA-dependent activation of the terminal complex of selenocysteine synthesis in humans.
Nucleic Acids Res., 51, 2023
|
|
7MDL
 
 | | High-resolution crystal structure of human SepSecS-tRNASec complex | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, CITRATE ANION, O-phosphoseryl-tRNA(Sec) selenium transferase, ... | | Authors: | Puppala, A, French, R.L, Simonovic, M. | | Deposit date: | 2021-04-05 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural basis for the tRNA-dependent activation of the terminal complex of selenocysteine synthesis in humans.
Nucleic Acids Res., 2023
|
|
7L1T
 
 | | Crystal structure of human holo SepSecS | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, O-phosphoseryl-tRNA(Sec) selenium transferase, PHOSPHATE ION | | Authors: | Puppala, A, Castillo Suchkou, J, Simonovic, M. | | Deposit date: | 2020-12-15 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the tRNA-dependent activation of the terminal complex of selenocysteine synthesis in humans.
Nucleic Acids Res., 2023
|
|
2R7B
 
 | | Crystal Structure of the Phosphoinositide-dependent Kinase-1 (PDK-1)Catalytic Domain bound to a dibenzonaphthyridine inhibitor | | Descriptor: | 10,11-dimethoxy-4-methyldibenzo[c,f]-2,7-naphthyridine-3,6-diamine, 3-phosphoinositide-dependent protein kinase 1, SULFATE ION | | Authors: | Olland, A.M. | | Deposit date: | 2007-09-07 | | Release date: | 2008-09-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Dibenzo[c,f][2,7]naphthyridines as Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 Inhibitors.
J.Med.Chem., 50, 2007
|
|
7Q6H
 
 | |
7Q7K
 
 | | JAK2 in complex with 4-(2-amino-8-methoxyquinazolin-6-yl)phenol | | Descriptor: | 4-(2-azanyl-8-methoxy-quinazolin-6-yl)phenol, Tyrosine-protein kinase JAK2 | | Authors: | Rowland, P. | | Deposit date: | 2021-11-09 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Investigation of Janus Kinase (JAK) Inhibitors for Lung Delivery and the Importance of Aldehyde Oxidase Metabolism.
J.Med.Chem., 65, 2022
|
|
7Q7L
 
 | |
7Q7I
 
 | |
7Q7W
 
 | |
3BCE
 
 | | Crystal structure of the ErbB4 kinase | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, Receptor tyrosine-protein kinase erbB-4, ... | | Authors: | Qiu, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Activation and Inhibition of the HER4/ErbB4 Kinase.
Structure, 16, 2008
|
|
