6R1X
 
 | |
6R1C
 
 | |
7R2Y
 
 | | Carbon regulatory PII-like protein SbtB from Synechocystis sp. 6803 in complex with ATP resulting from short ATP soak, conflicting T-loop and C-loop with partial occupancy | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Membrane-associated protein slr1513, SODIUM ION | | Authors: | Selim, K.A, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Carbon signaling protein SbtB possesses atypical redox-regulated apyrase activity to facilitate regulation of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7R30
 
 | | Carbon regulatory PII-like protein SbtB from Synechocystis sp. 6803 in complex with ADP and AMP resulting from ADP soak | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BICARBONATE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Selim, K.A, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carbon signaling protein SbtB possesses atypical redox-regulated apyrase activity to facilitate regulation of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7R2Z
 
 | |
7R32
 
 | | Carbon regulatory PII-like protein SbtB from Synechocystis sp. 6803, delta104 variant, in complex with ADP (co-crystal), tetragonal crystal form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Membrane-associated protein slr1513 | | Authors: | Selim, K.A, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Carbon signaling protein SbtB possesses atypical redox-regulated apyrase activity to facilitate regulation of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7R31
 
 | | Carbon regulatory PII-like protein SbtB from Synechocystis sp. 6803, C105A+C110A variant, in complex with ATP (co-crystal), tetragonal crystal form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Membrane-associated protein slr1513, ... | | Authors: | Selim, K.A, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Carbon signaling protein SbtB possesses atypical redox-regulated apyrase activity to facilitate regulation of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2WFW
 
 | | Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases - The Arc domain structure | | Descriptor: | ARC | | Authors: | Djuranovic, S, Hartmann, M.D, Habeck, M, Ursinus, A, Zwickl, P, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2009-04-15 | | Release date: | 2009-05-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
8RDS
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8i | | Descriptor: | (5~{S})-3-(2-methoxyphenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
8RDQ
 
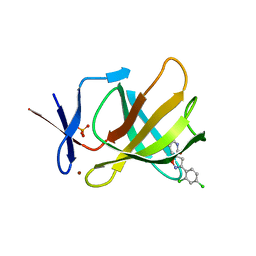 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8b | | Descriptor: | (5S)-3-(2,4-dichlorophenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
8RDP
 
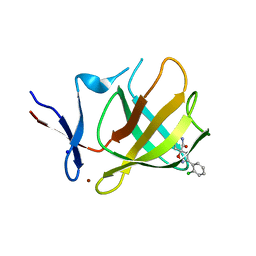 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8a | | Descriptor: | (5~{S})-3-(2-chlorophenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
8RDR
 
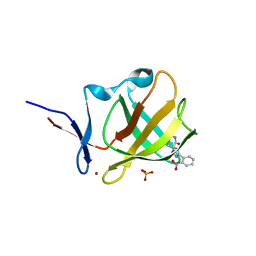 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8g | | Descriptor: | (5~{S})-3-(2-nitrophenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
2VC8
 
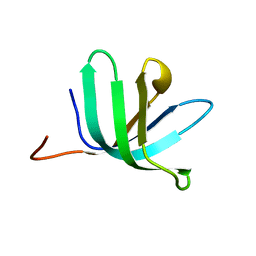 | |
8RDT
 
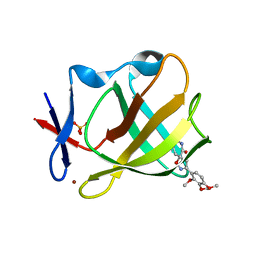 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8j | | Descriptor: | (5~{S})-3-(2,3,4-trimethoxyphenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
8OKH
 
 | |
8TLY
 
 | |
8TUK
 
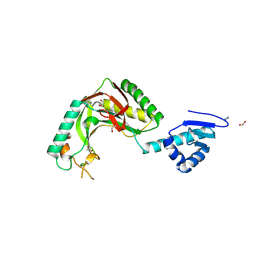 | | Alvinella ASCC1 KH and Phosphodiesterase/Ligase Domain | | Descriptor: | 1,2-ETHANEDIOL, Activating signal cointegrator 1 complex subunit 1, IMIDAZOLE | | Authors: | Tsutakawa, S.E, Tainer, J.A, Arvai, A.S, Chinnam, N.B. | | Deposit date: | 2023-08-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | ASCC1 structures and bioinformatics reveal a novel helix-clasp-helix RNA-binding motif linked to a two-histidine phosphodiesterase.
J.Biol.Chem., 300, 2024
|
|
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
2XQH
 
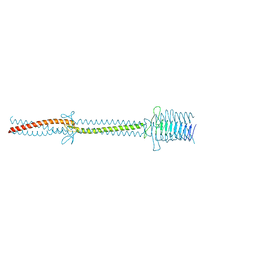 | | Crystal structure of an immunoglobulin-binding fragment of the trimeric autotransporter adhesin EibD | | Descriptor: | CHLORIDE ION, IMMUNOGLOBULIN-BINDING PROTEIN EIBD | | Authors: | Leo, J.C, Lyskowski, A, Hartmann, M, Schwarz, H, Linke, D, Lupas, A.N, Goldman, A. | | Deposit date: | 2010-09-02 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Structure of E. Coli Igg-Binding Protein D Suggests a General Model for Bending and Binding in Trimeric Autotransporter Adhesins.
Structure, 19, 2011
|
|
6SDA
 
 | | Bd2924 C10 acyl-coenzymeA bound form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable acyl-CoA dehydrogenase, decanoyl-CoA | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2019-07-26 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Target highlights in CASP13: Experimental target structures through the eyes of their authors.
Proteins, 87, 2019
|
|
6SD8
 
 | | Bd2924 apo-form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable acyl-CoA dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2019-07-26 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Target highlights in CASP13: Experimental target structures through the eyes of their authors.
Proteins, 87, 2019
|
|
7MHU
 
 | | Sialidase24 apo | | Descriptor: | Exo-alpha-sialidase | | Authors: | Rees, S.D, Chang, G.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational models in the service of X-ray and cryo-electron microscopy structure determination.
Proteins, 89, 2021
|
|
7M6B
 
 | | The Crystal Structure of Mcbe1 | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, S-ADENOSYLMETHIONINE, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2021-03-25 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Target highlights in CASP14: Analysis of models by structure providers.
Proteins, 89, 2021
|
|
7OC9
 
 | |
6GDX
 
 | |
