6CV0
 
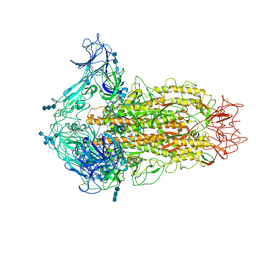 | | Cryo-electron microscopy structure of infectious bronchitis coronavirus spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shang, J, Zheng, Y, Yang, Y, Liu, C, Geng, Q, Luo, C, Zhang, W, Li, F. | | Deposit date: | 2018-03-27 | | Release date: | 2018-04-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins.
PLoS Pathog., 14, 2018
|
|
6BV0
 
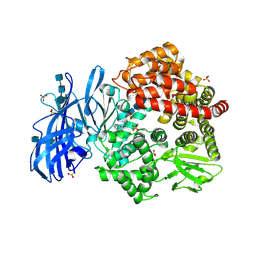 | | Crystal structure of porcine aminopeptidase-N with Arginine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
6BV2
 
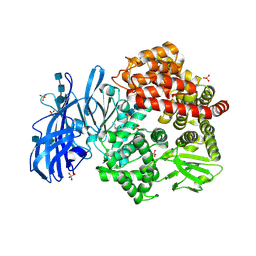 | | Crystal structure of porcine aminopeptidase-N with Isoleucine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
6BV1
 
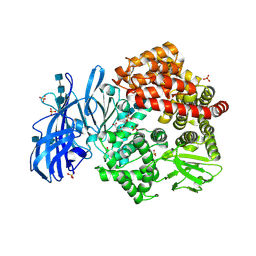 | | Crystal structure of porcine aminopeptidase-N with Aspartic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
6BV3
 
 | | Crystal structure of porcine aminopeptidase-N with Leucine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
8GY6
 
 | |
2LS2
 
 | |
2LS4
 
 | |
3CZ3
 
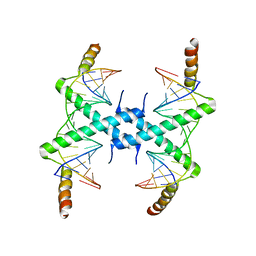 | | Crystal structure of Tomato Aspermy Virus 2b in complex with siRNA | | Descriptor: | Protein 2b, RNA (5'-R(P*CP*GP*UP*AP*CP*GP*CP*GP*GP*AP*AP*UP*AP*CP*UP*UP*CP*GP*A)-3'), RNA (5'-R(P*UP*CP*GP*AP*AP*GP*UP*AP*UP*UP*CP*CP*GP*CP*GP*UP*AP*CP*G)-3') | | Authors: | Ma, J.B, Li, F, Ding, S.W, Patel, D.J. | | Deposit date: | 2008-04-27 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structural Basis for siRNA Recognition by 2b, a Viral Suppressor of Non-Cell Autonomous RNA Silencing
To be Published
|
|
8T5I
 
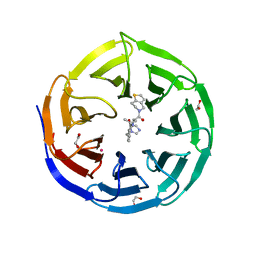 | | Crystal structure of human WDR5 in complex with MR4397 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, N-[(2S)-1-(6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl)-1-oxopentan-2-yl]-3-[(1H-imidazol-1-yl)methyl]benzamide, ... | | Authors: | Kimani, S, Dong, A, Li, F, Loppnau, P, Ackloo, S, Vedadi, M, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-13 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human WDR5 in complex with MR4397
To be published
|
|
1R8A
 
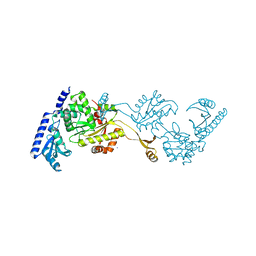 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide Complexes | | Descriptor: | MANGANESE (II) ION, SODIUM ION, tRNA nucleotidyltransferase | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
1R8B
 
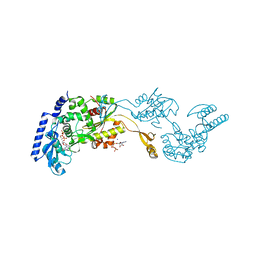 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | Descriptor: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKK
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody Nb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKL
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
5CQR
 
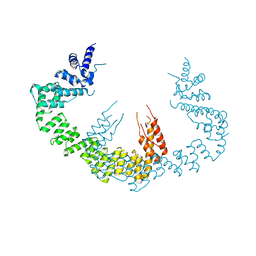 | | Dimerization of Elp1 is essential for Elongator complex assembly | | Descriptor: | Elongator complex protein 1 | | Authors: | Lin, Z, Xu, H, Li, F, Diao, W, Long, J, Shen, Y. | | Deposit date: | 2015-07-22 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.015 Å) | | Cite: | Dimerization of elongator protein 1 is essential for Elongator complex assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CQS
 
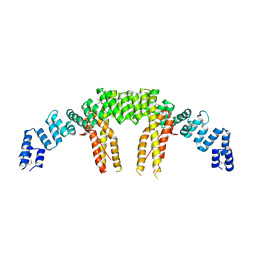 | | Dimerization of Elp1 is essential for Elongator complex assembly | | Descriptor: | Elongator complex protein 1 | | Authors: | Lin, Z, Xu, H, Li, F, Diao, W, Long, J, Shen, Y. | | Deposit date: | 2015-07-22 | | Release date: | 2015-08-19 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dimerization of elongator protein 1 is essential for Elongator complex assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5GJU
 
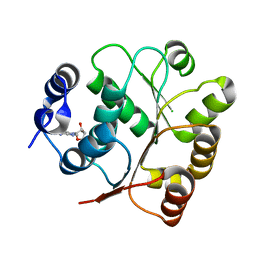 | | DEAD-box RNA helicase | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-dependent RNA helicase DeaD | | Authors: | Xu, L, Li, F, Wang, L, Shi, Y. | | Deposit date: | 2016-07-02 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
5GI4
 
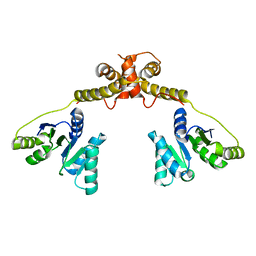 | | DEAD-box RNA helicase | | Descriptor: | ATP-dependent RNA helicase DeaD | | Authors: | Xu, L, Wang, L, Li, F, Wu, L, Shi, Y. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
5Y1U
 
 | | Crystal structure of RBBP4 bound to AEBP2 RRK motif | | Descriptor: | Histone-binding protein RBBP4, SULFATE ION, Zinc finger protein AEBP2 | | Authors: | Sun, A, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2017-07-21 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural and biochemical insights into human zinc finger protein AEBP2 reveals interactions with RBBP4
Protein Cell, 2017
|
|
5GMV
 
 | | LC3B-FUNDC1 complex | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, Peptide from FUN14 domain-containing protein 1 | | Authors: | Lv, M, Wang, C, Li, F. | | Deposit date: | 2016-07-17 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the recognition of phosphorylated FUNDC1 by LC3B in mitophagy
Protein Cell, 8, 2017
|
|
5Z4D
 
 | | Structure of Tailor in complex with AGUU RNA | | Descriptor: | RNA (5'-R(*AP*GP*UP*U)-3'), Terminal uridylyltransferase Tailor | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-11 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
5YYZ
 
 | | Crystal structure of the MEK1 FHA domain in complex with the HOP1 pThr318 peptide. | | Descriptor: | Meiosis-specific protein HOP1, Meiosis-specific serine/threonine-protein kinase MEK1 | | Authors: | Xie, C, Li, F, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-12-11 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural insights into the recognition of phosphorylated Hop1 by Mek1
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5Z4M
 
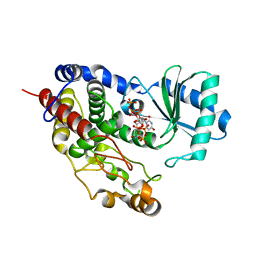 | | Structure of TailorD343A with bound UTP and Mg | | Descriptor: | MAGNESIUM ION, Terminal uridylyltransferase Tailor, URIDINE 5'-TRIPHOSPHATE | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-11 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
5Z4C
 
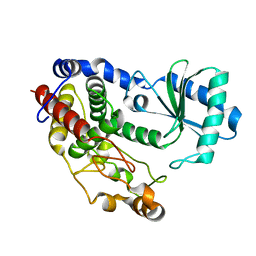 | | Crystal structure of Tailor | | Descriptor: | Terminal uridylyltransferase Tailor | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-10 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
