4CM5
 
 | |
4CM7
 
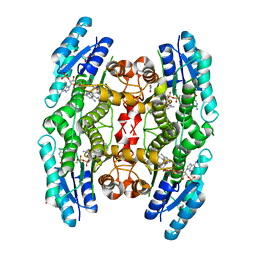 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | 5-phenethyl-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4CLE
 
 | |
4CZ1
 
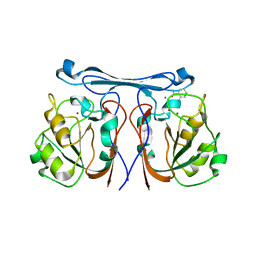 | | Crystal structure of kynurenine formamidase from Bacillus anthracis complexed with 2-aminoacetophenone. | | Descriptor: | 2-aminoacetophenone, KYNURENINE FORMAMIDASE, MAGNESIUM ION, ... | | Authors: | Diaz-Saez, L, Srikannathasan, V, Zoltner, M, Hunter, W.N. | | Deposit date: | 2014-04-16 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure of Bacterial Kynurenine Formamidase Reveals a Crowded Binuclear-Zinc Catalytic Site Primed to Generate a Potent Nucleophile.
Biochem.J., 462, 2014
|
|
1GY8
 
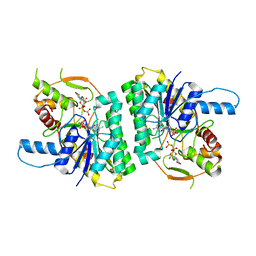 | | Trypanosoma brucei UDP-galactose 4' epimerase | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-GALACTOSE 4-EPIMERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Shaw, M.P, Bond, C.S, Hunter, W.N. | | Deposit date: | 2002-04-21 | | Release date: | 2003-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-Resolution Crystal Structure of Trypanosoma Brucei Udp-Galactose 4'-Epimerase: A Potential Target for Structure-Based Development of Novel Trypanocides
Mol.Biochem.Parasitol., 126, 2003
|
|
4CO9
 
 | | Crystal structure of kynurenine formamidase from Bacillus anthracis | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, KYNURENINE FORMAMIDASE, MAGNESIUM ION, ... | | Authors: | Diaz-Saez, L, Srikannathasan, V, Zoltner, M, Hunter, W.N. | | Deposit date: | 2014-01-27 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Bacterial Kynurenine Formamidase Reveals a Crowded Binuclear-Zinc Catalytic Site Primed to Generate a Potent Nucleophile.
Biochem.J., 462, 2014
|
|
1GXF
 
 | | CRYSTAL STRUCTURE OF TRYPANOSOMA CRUZI TRYPANOTHIONE REDUCTASE IN COMPLEX WITH THE INHIBITOR QUINACRINE MUSTARD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MALEIC ACID, QUINACRINE MUSTARD, ... | | Authors: | Bond, C.S, Peterson, M.R, Vickers, T.J, Fairlamb, A.H, Hunter, W.N. | | Deposit date: | 2002-04-04 | | Release date: | 2004-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two Interacting Binding Sites for Quinacrine Derivatives in the Active Site of Trypanothione Reductase: A Template for Drug Design
J.Biol.Chem., 279, 2004
|
|
4COG
 
 | | Crystal structure of kynurenine formamidase from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Diaz-Saez, L, Srikannathasan, V, Zoltner, M, Hunter, W.N. | | Deposit date: | 2014-01-28 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Bacterial Kynurenine Formamidase Reveals a Crowded Binuclear-Zinc Catalytic Site Primed to Generate a Potent Nucleophile.
Biochem.J., 462, 2014
|
|
4CM6
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | (E)-2-amino-4-oxo-6-styryl-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4CMK
 
 | |
4CMA
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | 5,6-diphenyl-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
1GUN
 
 | |
4CQI
 
 | | Crystal structure of recombinant tubulin-binding cofactor A (TBCA) from Leishmania major | | Descriptor: | GLYCEROL, SULFATE ION, TUBULIN-BINDING COFACTOR A | | Authors: | Barrack, K.L, Fyfe, P.K, Hunter, W.N. | | Deposit date: | 2014-02-17 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of Tubulin-Binding Cofactor a from Leishmania Major Infers a Mode of Association During the Early Stages of Microtubule Assembly
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4CM3
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 5-phenyl-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, ACETATE ION, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4CL8
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | 2,4-diamino-6-(3-formylphenyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4CMI
 
 | |
4CM4
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | 5-(4-fluorophenyl)-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, ACETATE ION, GLYCEROL, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4CLO
 
 | |
1GVF
 
 | | Structure of tagatose-1,6-bisphosphate aldolase | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOGLYCOLOHYDROXAMIC ACID, SODIUM ION, ... | | Authors: | Hall, D.R, Hunter, W.N. | | Deposit date: | 2002-02-11 | | Release date: | 2002-06-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of Tagatose-1,6-Bisphosphate Aldolase; Insight Into Chiral Discrimination, Mechanism and Specificity of Class II Aldolases
J.Biol.Chem., 277, 2002
|
|
1GX1
 
 | | Structure of 2C-Methyl-D-erythritol-2,4-cyclodiphosphate Synthase | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE SYNTHASE, CYTIDINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Kemp, L.E, Bond, C.S, Hunter, W.N. | | Deposit date: | 2002-03-26 | | Release date: | 2002-04-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of 2C-Methyl-D-Erythritol 2,4-Cyclodiphosphate Synthase: An Essential Enzyme for Isoprenoid Biosynthesis and Target for Antimicrobial Drug Development
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1H48
 
 | | The structure of 2C-Methyl-D-erythritol 2,4-cyclodiphosphate synthase in complex with CMP and product | | Descriptor: | 2C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE, 2C-METHYL-D-ERYTHRITOL-2,4-CYCLODIPHOSPHATE SYNTHASE, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Kemp, L.E, Alphey, M.S, Bond, C.S, Hunter, W.N. | | Deposit date: | 2003-02-24 | | Release date: | 2004-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Identification of Isoprenoids that Bind in the Intersubunit Cavity of Escherichia Coli 2C-Methyl-D-Erythritol-2,4-Cyclodiphosphate Synthase by Complementary Biophysical Methods
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1H9R
 
 | | Tungstate bound complex Dimop domain of ModE from E.coli | | Descriptor: | MOLYBDENUM TRANSPORT PROTEIN MODE, NICKEL (II) ION, TUNGSTATE(VI)ION | | Authors: | Gourley, D.G, Hunter, W.N. | | Deposit date: | 2001-03-19 | | Release date: | 2001-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Oxyanion Binding Alters Conformational and Quaternary Structure of the C-Terminal Domain of the Transcriptional Regulator Mode; Implications for Molybdate-Dependant Regulation, Signalling, Storage and Transport
J.Biol.Chem., 276, 2001
|
|
1H3M
 
 | | Structure of 4-diphosphocytidyl-2C-methyl-D-erythritol synthetase | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, CHLORIDE ION, PENTANE-1,5-DIAMINE | | Authors: | Kemp, L.E, Bond, C.S, Hunter, W.N. | | Deposit date: | 2002-09-10 | | Release date: | 2003-08-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Tetragonal Crystal Form of Escherichia Coli 2-C-Methyl-D-Erythritol 4-Phosphate Cytidylyltransferase
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1GUO
 
 | |
1GYN
 
 | | Class II fructose 1,6-bisphosphate aldolase with Cadmium (not Zinc) in the active site | | Descriptor: | CADMIUM ION, FRUCTOSE-BISPHOSPHATE ALDOLASE II | | Authors: | Hall, D.R, Kemp, L.E, Leonard, G.A, Berry, A, Hunter, W.N. | | Deposit date: | 2002-04-27 | | Release date: | 2003-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Organization of Divalent Cations in the Active Site of Cadmium Escherichia Coli Fructose 1,6-Bisphosphate Aldolase
Acta Crystallogr.,Sect.D, 59, 2003
|
|
