2FE9
 
 | |
8V2X
 
 | |
8V09
 
 | | Crystal Structure of the reconstruction of the ancestral triosephosphate isomerase of the last opisthokont common ancestor obtained by bayesian inference with PGH | | Descriptor: | ACETIC ACID, FORMIC ACID, PHOSPHOGLYCOLOHYDROXAMIC ACID, ... | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra-Borrego, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
8V2W
 
 | | Crystal Structure of the ancestral triosephosphate isomerase reconstruction of the last opisthokont common ancestor obtained by Bayesian inference | | Descriptor: | GLYCEROL, Triosephosphate isomerase | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
8TCO
 
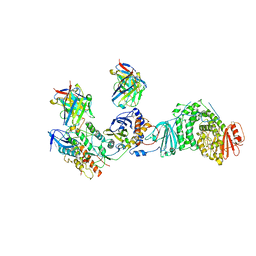 | | HCMV Trimer in complex with CS2it1p2_F7K Fab and CS4tt1p1_E3K Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CS2it1p2_F7K Fab heavy chain, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2023-07-02 | | Release date: | 2023-08-09 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Single-cell analysis of memory B cells from top neutralizers reveals multiple sites of vulnerability within HCMV Trimer and Pentamer.
Immunity, 56, 2023
|
|
2CW6
 
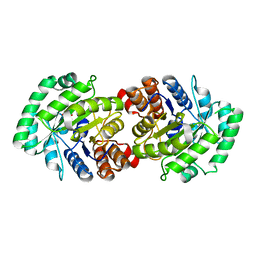 | | Crystal Structure of Human HMG-CoA Lyase: Insights into Catalysis and the Molecular Basis for Hydroxymethylglutaric Aciduria | | Descriptor: | 3-HYDROXYPENTANEDIOIC ACID, Hydroxymethylglutaryl-CoA lyase, mitochondrial, ... | | Authors: | Fu, Z, Runquist, J.A, Hunt, J.F, Miziorko, H.M, Kim, J.-J.P. | | Deposit date: | 2005-06-17 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human 3-hydroxy-3-methylglutaryl-CoA Lyase: insights into catalysis and the molecular basis for hydroxymethylglutaric aciduria
J.Biol.Chem., 281, 2006
|
|
8T5X
 
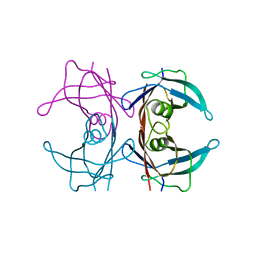 | | Probing the dissociation pathway of a kinetically labile transthyretin mutant (A25T) | | Descriptor: | Transthyretin | | Authors: | Ferguson, J.A, Sun, X, Leach, B.I, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2023-06-14 | | Release date: | 2023-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Probing the Dissociation Pathway of a Kinetically Labile Transthyretin Mutant.
J.Am.Chem.Soc., 146, 2024
|
|
6PRU
 
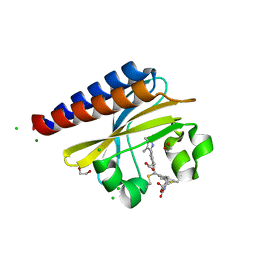 | | Photoconvertible crystals of PixJ from Thermosynechococcus elongatus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Burgie, E.S, Clinger, J.A, Miller, M.D, Phillips Jr, G.N, Vierstra, R.D. | | Deposit date: | 2019-07-11 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8SPK
 
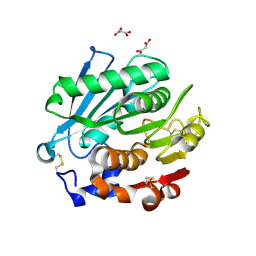 | | Crystal structure of Antarctic PET-degrading enzyme | | Descriptor: | Lipase 1, MALONATE ION | | Authors: | Furtado, A.A, Blazquez-Sanchez, P, Grinen, A, Vargas, J.A, Leonardo, D.A, Sculaccio, S.A, Pereira, H.M, Diez, B, Garratt, R.C, Ramirez-Sarmiento, C.A. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering the catalytic activity of an Antarctic PET-degrading enzyme by loop exchange.
Protein Sci., 32, 2023
|
|
6PQK
 
 | |
8V0A
 
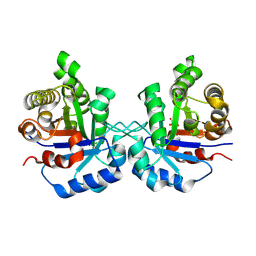 | | Crystal Structure of the worst case of the reconstruction of the ancestral triosephosphate isomerase of the last opisthokont common ancestor obtained by maximum likelihood with PGH | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Triosephosphate isomerase | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra-Borrego, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
6P7N
 
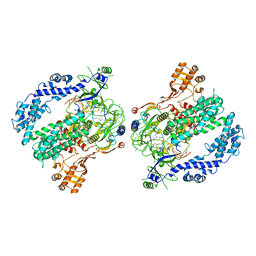 | | Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (2:2 complex) | | Descriptor: | Cas12a, MAGNESIUM ION, anti-CRISPR VA4, ... | | Authors: | Knott, G.J, Liu, J.J, Doudna, J.A. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-21 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis for AcrVA4 inhibition of specific CRISPR-Cas12a.
Elife, 8, 2019
|
|
2FQQ
 
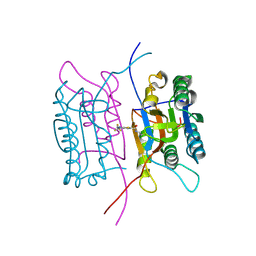 | | Crystal structure of human caspase-1 (Cys285->Ala, Cys362->Ala, Cys364->Ala, Cys397->Ala) in complex with 1-methyl-3-trifluoromethyl-1H-thieno[2,3-c]pyrazole-5-carboxylic acid (2-mercapto-ethyl)-amide | | Descriptor: | 1-METHYL-3-TRIFLUOROMETHYL-1H-THIENO[2,3-C]PYRAZOLE-5-CARBOXYLIC ACID (2-MERCAPTO-ETHYL)-AMIDE, Caspase-1 | | Authors: | Scheer, J.M, Wells, J.A, Romanowski, M.J. | | Deposit date: | 2006-01-18 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A common allosteric site and mechanism in caspases
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2FTC
 
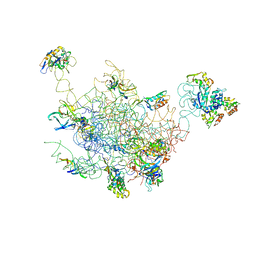 | | Structural Model for the Large Subunit of the Mammalian Mitochondrial Ribosome | | Descriptor: | 39S ribosomal protein L11, mitochondrial, 39S ribosomal protein L12, ... | | Authors: | Mears, J.A, Sharma, M.R, Gutell, R.R, Richardson, P.E, Agrawal, R.K, Harvey, S.C. | | Deposit date: | 2006-01-24 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.1 Å) | | Cite: | A Structural Model for the Large Subunit of the Mammalian Mitochondrial Ribosome
J.Mol.Biol., 358, 2006
|
|
2FWQ
 
 | |
3N5F
 
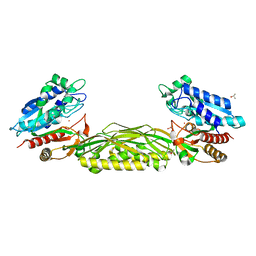 | | Crystal Structure of L-N-carbamoylase from Geobacillus stearothermophilus CECT43 | | Descriptor: | CACODYLATE ION, COBALT (II) ION, ISOPROPYL ALCOHOL, ... | | Authors: | Garcia-Pino, A, Martinez-Rodriguez, S, Gavira, J.A. | | Deposit date: | 2010-05-25 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mutational and structural analysis of L-N-carbamoylase reveals new insights into a peptidase m20/m25/m40 family member.
J.Bacteriol., 194, 2012
|
|
2FFL
 
 | |
2FLJ
 
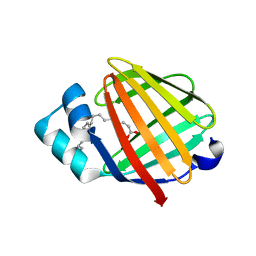 | | Fatty acid binding protein from locust flight muscle in complex with oleate | | Descriptor: | Fatty acid-binding protein, OLEIC ACID | | Authors: | Luecke, C, Qiao, Y, van Moerkerk, H.T.B, Veerkamp, J.H, Hamilton, J.A. | | Deposit date: | 2006-01-06 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Fatty-acid-binding protein from the flight muscle of Locusta migratoria: evolutionary variations in fatty acid binding.
Biochemistry, 45, 2006
|
|
8UVT
 
 | |
2FAT
 
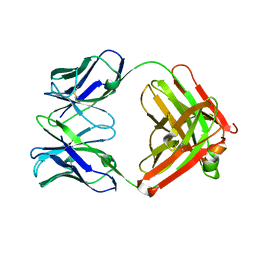 | | An anti-urokinase plasminogen activator receptor (UPAR) antibody: Crystal structure and binding epitope | | Descriptor: | FAB ATN-615, heavy chain, light chain | | Authors: | Li, Y, Parry, G, Shi, X, Chen, L, Callahan, J.A, Mazar, A.P, Huang, M. | | Deposit date: | 2005-12-07 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | An anti-urokinase plasminogen activator receptor (uPAR) antibody: crystal structure and binding epitope
J.Mol.Biol., 365, 2007
|
|
2FCH
 
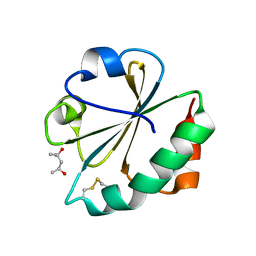 | |
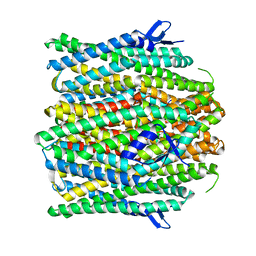
8UVU
 
 | |
2FSY
 
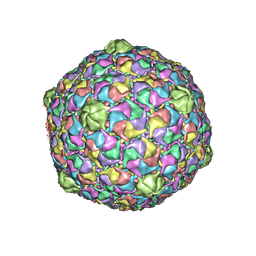 | | Bacteriophage HK97 Pepsin-treated Expansion Intermediate IV | | Descriptor: | major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-23 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
3N37
 
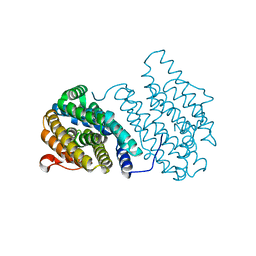 | | Ribonucleotide Reductase Dimanganese(II)-NrdF from Escherichia coli | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Ribonucleoside-diphosphate reductase 2 subunit beta | | Authors: | Boal, A.K, Cotruvo Jr, J.A, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2010-05-19 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for activation of class Ib ribonucleotide reductase.
Science, 329, 2010
|
|
2FZL
 
 | |
