6WGR
 
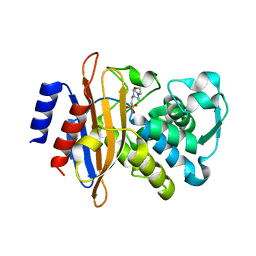 | | The crystal structure of a beta-lactamase from Staphylococcus aureus subsp. aureus USA300_TCH1516 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase, GLYCEROL | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-06 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The crystal structure of a beta-lactamase from Staphylococcus aureus subsp. aureus USA300_TCH1516
To Be Published
|
|
1R58
 
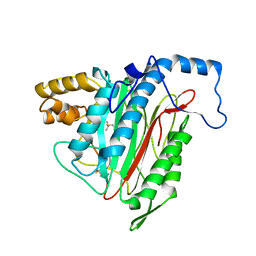 | | Crystal Structure of MetAP2 complexed with A357300 | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2, N'-((2S,3R)-3-AMINO-2-HYDROXY-5-(ISOPROPYLSULFANYL)PENTANOYL)-N-3-CHLOROBENZOYL HYDRAZIDE | | Authors: | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Ericken, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
6X5P
 
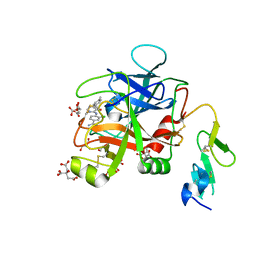 | | Discovery of Hydroxy Pyrimidine Factor IXa Inhibitors | | Descriptor: | 3-chloro-4-{[5-hydroxy-6-(4-methylphenyl)pyrimidin-4-yl]amino}benzene-1-carboximidamide, CITRIC ACID, Coagulation factor IX, ... | | Authors: | Jayne, C.L, Andreani, T, Chan, T, Chelliah, M.V, Clasby, M.C, Dwyer, M, Eagen, K.A, Fried, S, Greenlee, W.J, Guo, Z, Hawes, B, Hruza, A, Ingram, R, Keertikar, K.M, Neelamkavil, S, Reichert, P, Xia, Y, Chackalamannil, S. | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Discovery of hydroxy pyrimidine Factor IXa inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
2AWV
 
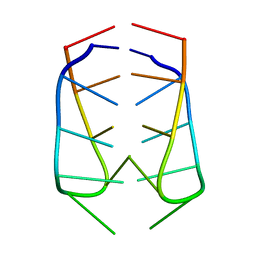 | | NMR Structural Analysis of the dimer of 5MCCTCATCC | | Descriptor: | 5'-D(*(MCY)P*CP*TP*CP*AP*CP*TP*CP*C)-3' | | Authors: | Canalia, M, Leroy, J.-L. | | Deposit date: | 2005-09-02 | | Release date: | 2005-09-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, internal motions and association-dissociation kinetics of the i-motif dimer of d(5mCCTCACTCC).
Nucleic Acids Res., 33, 2005
|
|
1R7A
 
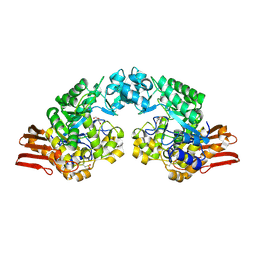 | | Sucrose Phosphorylase from Bifidobacterium adolescentis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, sucrose phosphorylase | | Authors: | Sprogoe, D, van den Broek, L.A.M, Mirza, O, Kastrup, J.S, Voragen, A.G.J, Gajhede, M, Skov, L.K. | | Deposit date: | 2003-10-21 | | Release date: | 2004-02-10 | | Last modified: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of sucrose phosphorylase from Bifidobacterium adolescentis.
Biochemistry, 43, 2004
|
|
1R7S
 
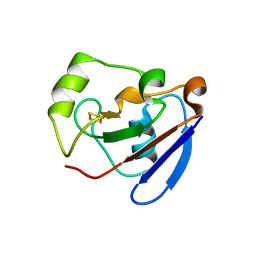 | | PUTIDAREDOXIN (Fe2S2 ferredoxin), C73G mutant | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Putidaredoxin | | Authors: | Smith, N, Mayhew, M, Kelly, H, Robinson, H, Heroux, A, Holden, M.J, Gallagher, D.T. | | Deposit date: | 2003-10-22 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of C73G putidaredoxin from Pseudomonas putida.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2AZD
 
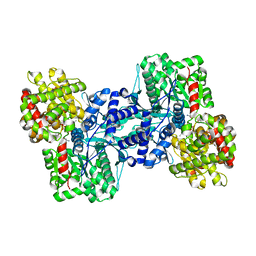 | |
6WML
 
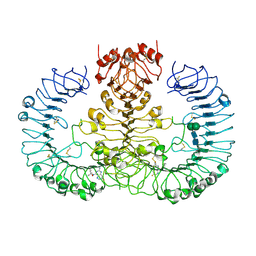 | | Human TLR8 bound to the potent agonist, GS-9688 (Selgantolimod) | | Descriptor: | (2R)-2-[(2-amino-7-fluoropyrido[3,2-d]pyrimidin-4-yl)amino]-2-methylhexan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 8, ... | | Authors: | Appleby, T.C, Perry, J.K, Mish, M, Villasenor, A.G, Mackman, R.L. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of GS-9688 (Selgantolimod) as a Potent and Selective Oral Toll-Like Receptor 8 Agonist for the Treatment of Chronic Hepatitis B.
J.Med.Chem., 63, 2020
|
|
6WNQ
 
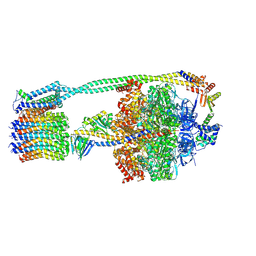 | | E. coli ATP Synthase State 2a | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
2ATV
 
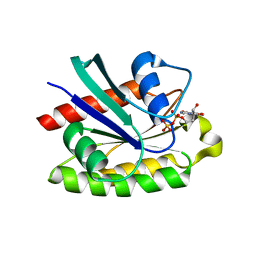 | | The crystal structure of human RERG in the GDP bound state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAS-like estrogen-regulated growth inhibitor | | Authors: | Turnbull, A.P, Salah, E, Schoch, G, Elkins, J, Burgess, N, Gileadi, O, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-26 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of human RERG in the GDP bound state
To be Published
|
|
2B5U
 
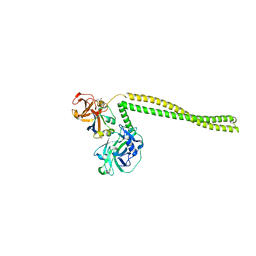 | | Crystal Structure Of Colicin E3 V206C Mutant In Complex With Its Immunity Protein | | Descriptor: | CITRIC ACID, Colicin E3, Colicin E3 immunity protein | | Authors: | Nallini Vijayarangan, A, Nithianantham, S, Nan, W, Jakes, K, Shoham, M. | | Deposit date: | 2005-09-29 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Of Colicin E3 In Complex With Its Immunity Protein
TO BE PUBLISHED
|
|
1RDW
 
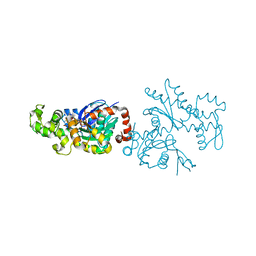 | | Actin Crystal Dynamics: Structural Implications for F-actin Nucleation, Polymerization and Branching Mediated by the Anti-parallel Dimer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reutzel, R, Yoshioka, C, Govindasamy, L, Yarmola, E.G, Agbandje-Mckenna, M, Bubb, M.R, Mckenna, R. | | Deposit date: | 2003-11-06 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Actin crystal dynamics: structural implications for F-actin nucleation, polymerization, and branching mediated by the anti-parallel dimer.
J.Struct.Biol., 146, 2004
|
|
6WVL
 
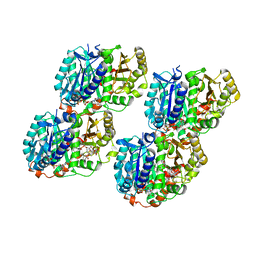 | | Low curvature lateral interaction within a 13-protofilament, Taxol stabilized microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Debs, G.E, Cha, M, Huehn, A.R, Sindelar, C.V. | | Deposit date: | 2020-05-06 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dynamic and asymmetric fluctuations in the microtubule wall captured by high-resolution cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2B8A
 
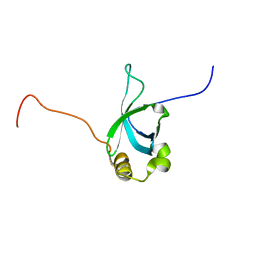 | | High Resolution Structure of the HDGF PWWP Domain | | Descriptor: | Hepatoma-derived growth factor | | Authors: | Lukasik, S.M, Cierpicki, T, Borloz, M, Grembecka, J, Everett, A, Bushweller, J.H. | | Deposit date: | 2005-10-06 | | Release date: | 2005-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution structure of the HDGF PWWP domain: a potential DNA binding domain.
Protein Sci., 15, 2006
|
|
1RHW
 
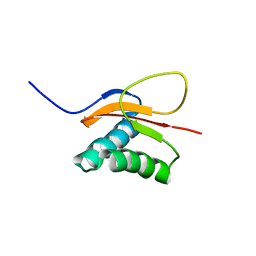 | | The solution structure of the pH-induced monomer of dynein light chain LC8 from Drosophila | | Descriptor: | Dynein light chain 1, cytoplasmic | | Authors: | Makokha, M, Huang, Y.J, Montelione, G, Edison, A.S, Barbar, E. | | Deposit date: | 2003-11-14 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the pH-induced monomer of dynein light-chain LC8 from Drosophila.
Protein Sci., 13, 2004
|
|
1RI7
 
 | |
6X0V
 
 | | Structure of MZT2/GCP-NHD and CDK5Rap2 at position 13 of the gamma-TuRC | | Descriptor: | Centrosome protein Cep215, Gamma-tubulin complex component 2, Mitotic-spindle organizing protein 2A | | Authors: | Wieczorek, M, Huang, T.-L, Urnavicius, L, Hsia, K.-C, Kapoor, T.M. | | Deposit date: | 2020-05-17 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | MZT Proteins Form Multi-Faceted Structural Modules in the gamma-Tubulin Ring Complex.
Cell Rep, 31, 2020
|
|
6WZ5
 
 | |
6X24
 
 | | Structural basis of plant blue light photoreceptor | | Descriptor: | Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Palayam, M, Ganapathy, J, Guercio, M.A, Shabek, N. | | Deposit date: | 2020-05-19 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural insights into photoactivation of plant Cryptochrome-2.
Commun Biol, 4, 2021
|
|
1RJF
 
 | | Structure of PPM1, a leucine carboxy methyltransferase involved in the regulation of protein phosphatase 2A activity | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, carboxy methyl transferase for protein phosphatase 2A catalytic subunit | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Sorel, I, de La Sierra-Gallay, I.L, Collinet, B, Graille, M, Blondeau, K, Bettache, N, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2003-11-19 | | Release date: | 2003-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of protein phosphatase methyltransferase 1 (PPM1), a leucine carboxyl methyltransferase involved in the regulation of protein phosphatase 2A activity
J.Biol.Chem., 279, 2004
|
|
2AHQ
 
 | |
1R1B
 
 | | EPRS SECOND REPEATED ELEMENT, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRNA SYNTHETASE | | Authors: | Cahuzac, B, Berthonneau, E, Birlirakis, N, Mirande, M, Guittet, E. | | Deposit date: | 1998-12-15 | | Release date: | 1999-12-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A recurrent RNA-binding domain is appended to eukaryotic aminoacyl-tRNA synthetases.
EMBO J., 19, 2000
|
|
1RKR
 
 | | CRYSTAL STRUCTURE OF AZURIN-I FROM ALCALIGENES XYLOSOXIDANS NCIMB 11015 | | Descriptor: | AZURIN-I, COPPER (II) ION | | Authors: | Li, C, Inoue, T, Gotowda, M, Suzuki, S, Yamaguchi, K, Kataoka, K, Kai, Y. | | Deposit date: | 1997-05-17 | | Release date: | 1998-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of azurin I from the denitrifying bacterium Alcaligenes xylosoxidans NCIMB 11015 at 2.45 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2ARP
 
 | | Activin A in complex with Fs12 fragment of follistatin | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Follistatin, GLYCEROL, ... | | Authors: | Harrington, A.E, Morris-Triggs, S.A, Ruotolo, B.T, Robinson, C.V, Ohnuma, S, Hyvonen, M. | | Deposit date: | 2005-08-21 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the inhibition of activin signalling by follistatin
Embo J., 25, 2006
|
|
2AS0
 
 | | Crystal Structure of PH1915 (APC 5817): A Hypothetical RNA Methyltransferase | | Descriptor: | hypothetical protein PH1915 | | Authors: | Sun, W, Xu, X, Pavlova, M, Edwards, A.M, Joachimiak, A, Savchenko, A, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-22 | | Release date: | 2005-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a novel SAM-dependent methyltransferase PH1915 from Pyrococcus horikoshii.
Protein Sci., 14, 2005
|
|
