4RHD
 
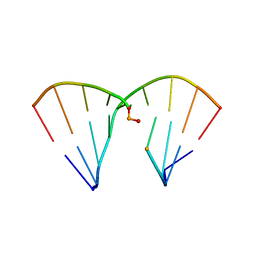 | | DNA Duplex with Novel ZP Base Pair | | Descriptor: | DNA 9mer novel P nucleobase, DNA 9mer novel Z nucleobase, MAGNESIUM ION | | Authors: | Zhang, W, Zhang, L, Benner, S, Huang, Z. | | Deposit date: | 2014-10-01 | | Release date: | 2015-07-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of functional six-nucleotide DNA.
J.Am.Chem.Soc., 137, 2015
|
|
6KZI
 
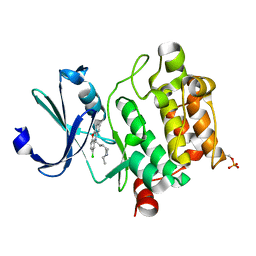 | | Crystal structure of Ser/Thr kinase Pim1 in complex with thioridazine derivatives | | Descriptor: | 4-(2-chloro-10H-phenoxazin-10-yl)-N,N-diethylbutan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Wan, X, Li, W, Xie, Y, Huang, N. | | Deposit date: | 2019-09-24 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
1U7R
 
 | |
1JXZ
 
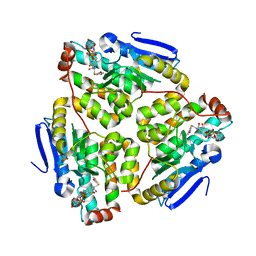 | | Structure of the H90Q mutant of 4-Chlorobenzoyl-Coenzyme A Dehalogenase complexed with 4-hydroxybenzoyl-Coenzyme A (product) | | Descriptor: | 4-HYDROXYBENZOYL COENZYME A, 4-chlorobenzoyl Coenzyme A dehalogenase, CALCIUM ION, ... | | Authors: | Thoden, J.B, Zhang, W, Wei, Y, Luo, L, Taylor, K.L, Yang, G, Dunaway-Mariano, D, Benning, M.M, Holden, H.M. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Histidine 90 Function in 4-chlorobenzoyl-coenzyme A Dehalogenase Catalysis
Biochemistry, 40, 2001
|
|
6L15
 
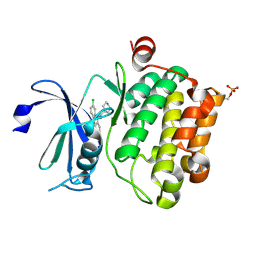 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 7-chloranyl-5-[3-[(3~{S})-piperidin-3-yl]propyl]pyrido[3,4-b][1,4]benzoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L12
 
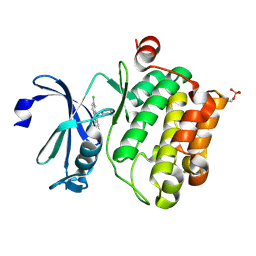 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 4-[(2-chloranylphenoxazin-10-yl)methyl]cyclohexan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L16
 
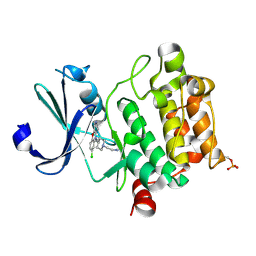 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-[4-[2-(7-chloranylpyrido[3,4-b][1,4]benzoxazin-5-yl)ethyl]piperidin-1-yl]ethanamine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L11
 
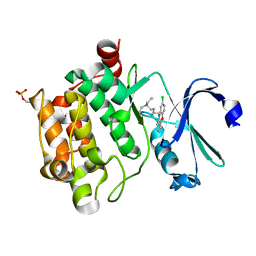 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 5-(2-chloranylphenoxazin-10-yl)-~{N},~{N}-diethyl-pentan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L17
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 7-chloranyl-5-[3-[(3~{S})-piperidin-3-yl]propyl]pyrido[3,4-b][1,4]benzoxazin-8-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L13
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-chloranyl-10-(2-piperidin-4-ylethyl)phenoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L14
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-chloranyl-10-[3-[(3~{S})-piperidin-3-yl]propyl]phenoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
4I41
 
 | | Crystal Structure of human Ser/Thr kinase Pim1 in complex with mitoxantrone | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Wan, X, Xie, Y, Huang, N. | | Deposit date: | 2012-11-27 | | Release date: | 2013-12-11 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new target for an old drug: identifying mitoxantrone as a nanomolar inhibitor of PIM1 kinase via kinome-wide selectivity modeling.
J. Med. Chem., 56, 2013
|
|
8SX6
 
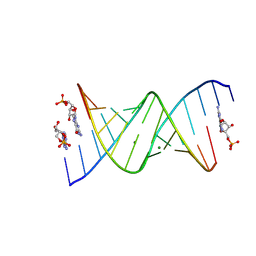 | | RNA duplex bound with GMP and AMP monomers | | Descriptor: | ADENOSINE MONOPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
8SWG
 
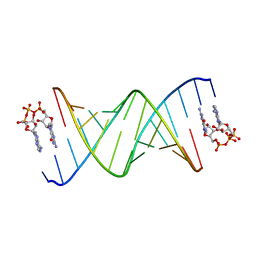 | | RNA duplex bound with GpppA dinucleotide ligand | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G*(GA3))-3') | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
8SX5
 
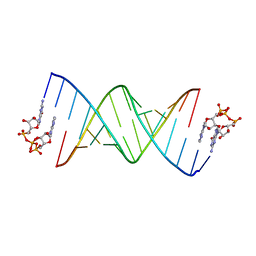 | | GpppA dinucleotide binding to RNA CU template | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA (5'-R(*(TLN)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G*(GA3))-3') | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
8SXL
 
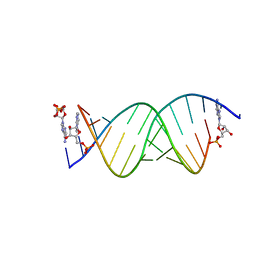 | | RNA UU template binding to AMP monomer | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA (5'-R(*(TLN)P*(TLN)P*(LCA)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-22 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
8SWO
 
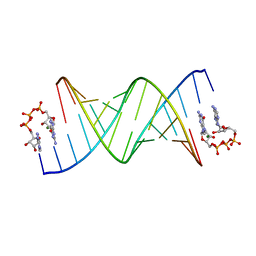 | | GpppA dinucleotide ligand binding to RNA UC template | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (5'-R(*(TLN)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G*(GA3))-3') | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
8SY1
 
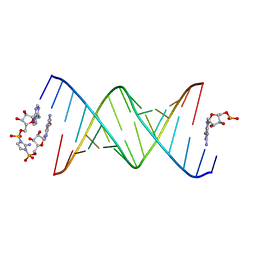 | | RNA duplex bound with imidazolium bridged GA dinucleotide | | Descriptor: | RNA (5'-R(*(TLN)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G*(GMA))-3'), [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-[2-azanyl-3-[[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]imidazol-1-yl]phosphinic acid | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
2L83
 
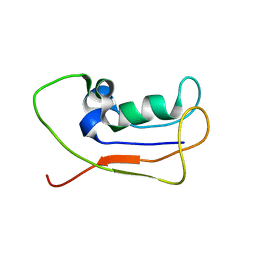 | | A protein from Haloferax volcanii | | Descriptor: | Small archaeal modifier protein 1 | | Authors: | Zhang, W, Liao, S, Fan, K, Tu, X. | | Deposit date: | 2011-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ionic strength-dependent conformations of a ubiquitin-like small archaeal modifier protein (SAMP1) from Haloferax volcanii.
Protein Sci., 22, 2013
|
|
6K0R
 
 | | Ruvbl1-Ruvbl2 with truncated domain II in complex with phosphorylated Cordycepin | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, W, Chen, L, Li, W, Ju, D, Huang, N, Zhang, E. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Chemical perturbations reveal that RUVBL2 regulates the circadian phase in mammals.
Sci Transl Med, 12, 2020
|
|
8F27
 
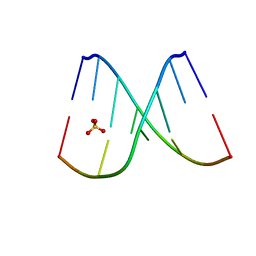 | | Mirror-image DNA containing 2'-OMe-L-uridine residue | | Descriptor: | Mirror-image DNA (5'-D(*(0DG)P*(0MU)P*(0DG)P*(0DT)P*(0DA)P*(0DC)P*(0DA)P*(0DC))-3'), SULFATE ION | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2022-11-07 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Derivatization of Mirror-Image l-Nucleic Acids with 2'-OMe Modification for Thermal and Structural Stabilization.
Chembiochem, 24, 2023
|
|
8F5C
 
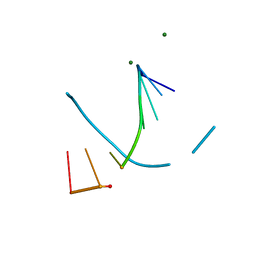 | | Mirror-image DNA containing 2'-OMe-L-dC modification | | Descriptor: | DNA (5'-D(*(0DG))-R(P*(XE6))-D(P*(0DG)P*(0DT)P*(0DA)P*(0DC)P*(0DG)P*(0DC))-3'), MAGNESIUM ION | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2022-11-13 | | Release date: | 2023-09-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Synthesis and Structural Characterization of 2'-Deoxy-2'-Methoxy-L-Cytidine Nucleic Acids
Chemistryselect, 2023
|
|
6U7Z
 
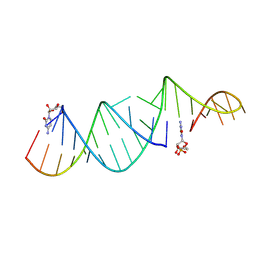 | | RNA hairpin structure containing one TNA nucleotide as primer | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, RNA (5'-R(*CP*UP*AP*UP*GP*CP*CP*UP*GP*CP*UP*G)-3'), RNA (5'-R(*CP*UP*GP*CP*UP*GP*GP*CP*UP*AP*AP*GP*GP*GP*CP*CP*GP*AP*AP*AP*GP*(TG))-3') | | Authors: | Szostak, J.W, Zhang, W. | | Deposit date: | 2019-09-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural interpretation of the effects of threo-nucleotides on nonenzymatic template-directed polymerization.
Nucleic Acids Res., 49, 2021
|
|
6U8U
 
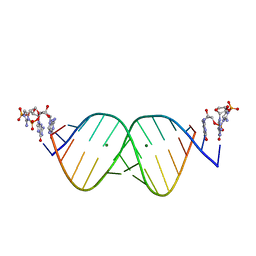 | | RNA duplex bound with TNA 3'-3' imidazolium dimer | | Descriptor: | 2-amino-3-[(R)-{[(3S,4R,5R)-5-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-4-hydroxyoxolan-3-yl]oxy}(hydroxy)phosphoryl]-1-[(S)-{[(3S,4R,5R)-5-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-4-hydroxyoxolan-3-yl]oxy}(hydroxy)phosphoryl]-1H-imidazol-3-ium, MAGNESIUM ION, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G)-3') | | Authors: | Szostak, J.W, Zhang, W. | | Deposit date: | 2019-09-05 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural interpretation of the effects of threo-nucleotides on nonenzymatic template-directed polymerization.
Nucleic Acids Res., 49, 2021
|
|
6U89
 
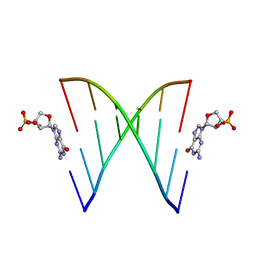 | | RNA duplex, bound with TNA monomer | | Descriptor: | 2-azanyl-9-[(2~{R},3~{R},4~{S})-3-oxidanyl-4-[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxy-oxolan-2-yl]-1~{H}-purin-6-one, RNA (5'-R(*(LCC)P*(TLN)P*(LCG)P*UP*AP*CP*A)-3') | | Authors: | Szostak, J.W, Zhang, W. | | Deposit date: | 2019-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural interpretation of the effects of threo-nucleotides on nonenzymatic template-directed polymerization.
Nucleic Acids Res., 49, 2021
|
|
