2KYI
 
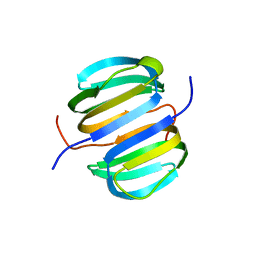 | | Solution NMR structure of Dsy0195(21-82) protein from Desulfitobacterium Hafniense. Northeast Structural Genomics Consortium Target DhR8C | | Descriptor: | Uncharacterized protein | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Wang, H, Ciccosanti, C, Foote, E.L, Jiang, M, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-05-27 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Combining NMR and EPR methods for homodimer protein structure determination.
J.Am.Chem.Soc., 132, 2010
|
|
2KIW
 
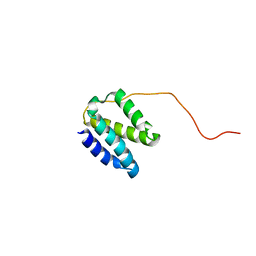 | | Solution NMR structure of the domain N-terminal to the integrase domain of SH1003 from Staphylococcus haemolyticus. Northeast Structural Genomics Consortium Target ShR105F (64-166). | | Descriptor: | Int protein | | Authors: | Yang, Y, Ramelot, T.A, Belote, R.L, Foote, E.L, Janjua, H, Nair, R, Rost, B, Swapna, G, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-12 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the domain N-terminal to the integrase domain of SH1003 from Staphylococcus
haemolyticus. Northeast Structural Genomics Consortium Target ShR105F (64-166).
To be Published
|
|
8H9H
 
 | | Crystal structure of ZBTB7A in complex with GACCC-containing sequence | | Descriptor: | DNA (5'-D(*TP*AP*AP*GP*GP*AP*CP*CP*CP*AP*GP*AP*T)-3'), DNA (5'-D(P*AP*AP*TP*CP*TP*GP*GP*GP*TP*CP*CP*TP*T)-3'), ZINC ION, ... | | Authors: | Yang, Y. | | Deposit date: | 2022-10-25 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ZBTB7A regulates primed-to-naive transition of pluripotent stem cells via recognition of the PNT-associated sequence by zinc fingers 1-2 and recognition of gamma-globin -200 gene element by zinc fingers 1-4.
Febs J., 290, 2023
|
|
6IS5
 
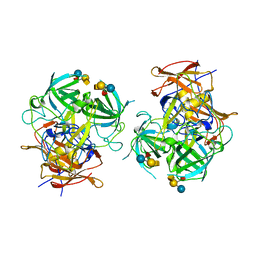 | | P domain of GII.3-TV24 with A-tetrasaccharide complex | | Descriptor: | VP1 Capsid protein, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yang, Y. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
6IR5
 
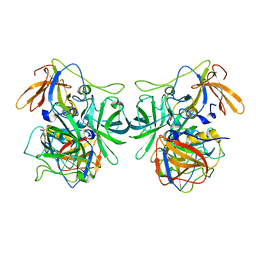 | | P domain of GII.3-TV24 | | Descriptor: | VP1 Capsid protein | | Authors: | Yang, Y. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
6KCZ
 
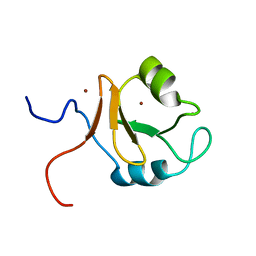 | |
6KZE
 
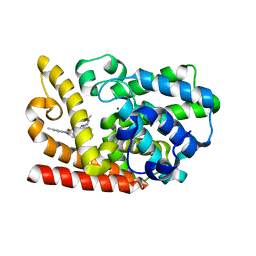 | | The crystal structue of PDE10A complexed with 4d | | Descriptor: | 8-[(E)-2-[5-methyl-1-[3-[3-(4-methylpiperazin-1-yl)propoxy]phenyl]benzimidazol-2-yl]ethenyl]quinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Yang, Y, Zhang, S, Zhou, Q, Huang, Y.-Y, Guo, L, Luo, H.-B. | | Deposit date: | 2019-09-24 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.50003481 Å) | | Cite: | Novel Potent and Highly Selective Benzoimidazole-based Phosphodiesterase 10 Inhibitors with Improved Solubility and Pharmacokinetic Properties for the Treatment of Pulmonary Arterial Hypertension
To Be Published
|
|
8TLV
 
 | |
8TLX
 
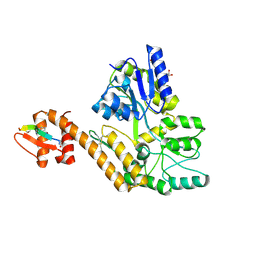 | |
8TLW
 
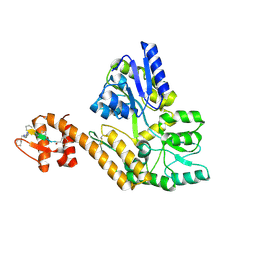 | |
8WS3
 
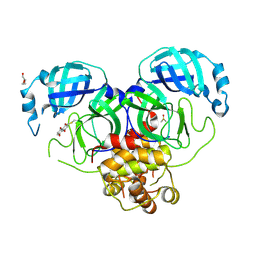 | | Crystal structure of SARS-CoV-2 Main Protease (Mpro) with covalent inhibitor 5,8-Dihydroxy-1,4-naphthoquinone | | Descriptor: | 3C-like proteinase nsp5, 5,8-bis(oxidanyl)naphthalene-1,4-dione, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, Y, Wu, D. | | Deposit date: | 2023-10-16 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Covalent inhibitors of SARS-CoV-2 main protease
To Be Published
|
|
6LGJ
 
 | | Crystal structure of an oxido-reductase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yang, Y, Lei, J, Yin, L. | | Deposit date: | 2019-12-05 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an oxido-reductase
To be published
|
|
6LGK
 
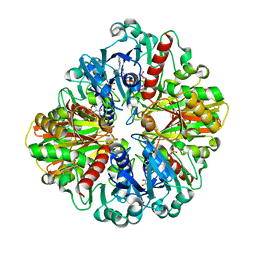 | |
2JOB
 
 | | Solution structure of an antilipopolysaccharide factor from shrimp and its possible Lipid A binding site | | Descriptor: | antilipopolysaccharide factor | | Authors: | Yang, Y, Boze, H, Chemardin, P, Padilla, A, Moulin, G, Tassanakajon, A, Pugniere, M, Roquet, F, Gueguen, Y, Bachere, E, Aumelas, A. | | Deposit date: | 2007-03-02 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR structure of rALF-Pm3, an anti-lipopolysaccharide factor from shrimp: Model of the possible lipid A-binding site
Biopolymers, 91, 2009
|
|
5ZTE
 
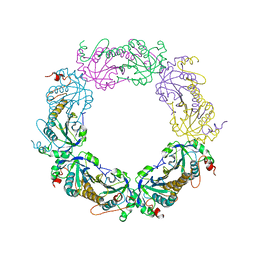 | | Crystal structure of PrxA C119S mutant from Arabidopsis thaliana | | Descriptor: | 2-Cys peroxiredoxin BAS1, chloroplastic | | Authors: | Yang, Y, Cai, W, Wang, J, Pan, W, Liu, L, Wang, M, Zhang, M. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Arabidopsis thaliana peroxiredoxin A C119S mutant.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7DUU
 
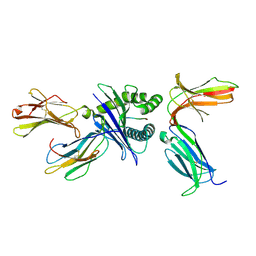 | | Crystal structure of HLA molecule with an KIR receptor | | Descriptor: | Beta-2-microglobulin, Killer cell immunoglobulin-like receptor 2DS2, LEU-ASN-PRO-SER-VAL-ALA-ALA-THR-LEU, ... | | Authors: | Yang, Y, Yin, L. | | Deposit date: | 2021-01-11 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Activating receptor KIR2DS2 bound to HLA-C1 reveals the novel recognition features of activating receptor.
Immunology, 165, 2022
|
|
3I5F
 
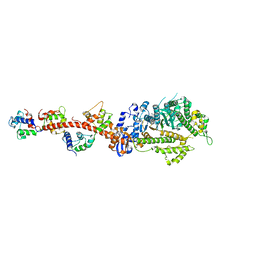 | | Crystal structure of squid MG.ADP myosin S1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin catalytic light chain LC-1, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2009-07-05 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
3I5G
 
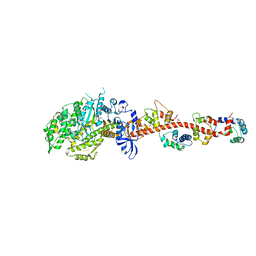 | | Crystal structure of rigor-like squid myosin S1 | | Descriptor: | CALCIUM ION, MALONATE ION, Myosin catalytic light chain LC-1, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2009-07-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
3I5I
 
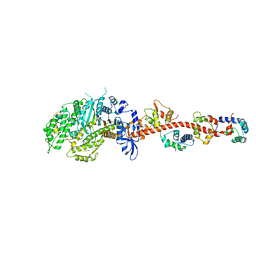 | | The crystal structure of squid myosin S1 in the presence of SO4 2- | | Descriptor: | CALCIUM ION, Myosin catalytic light chain LC-1, mantle muscle, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2009-07-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
3I5H
 
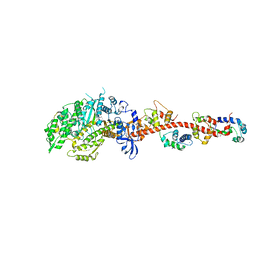 | | The crystal structure of rigor like squid myosin S1 in the absence of nucleotide | | Descriptor: | CALCIUM ION, Myosin catalytic light chain LC-1, mantle muscle, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2009-07-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
6COS
 
 | | CSP1-f11 | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6COT
 
 | | CSP2-d10 | | Descriptor: | Competence-stimulating peptide type 2 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2018-09-19 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6COO
 
 | | CSP1-E1A | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6COR
 
 | | CSP1-F11A | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6COU
 
 | | CSP2-E1Ad10 | | Descriptor: | Competence-stimulating peptide type 2 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2018-09-19 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
