2PZF
 
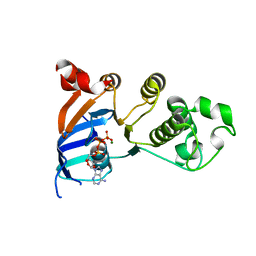 | | Minimal human CFTR first nucleotide binding domain as a head-to-tail dimer with delta F508 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Atwell, S, Conners, K, Emtage, S, Gheyi, T, Glenn, N.R, Hendle, J, Lewis, H.A, Lu, F, Rodgers, L.A, Romero, R, Sauder, J.M, Smith, D, Tien, H, Wasserman, S.R, Zhao, X. | | Deposit date: | 2007-05-18 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant.
Protein Eng.Des.Sel., 23, 2010
|
|
2N72
 
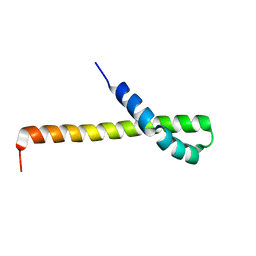 | | Solution structure of the Q domain from ACBD3 | | Descriptor: | Golgi resident protein GCP60 | | Authors: | Veverka, V, Hexnerova, R. | | Deposit date: | 2015-09-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights and in vitro reconstitution of membrane targeting and activation of human PI4KB by the ACBD3 protein.
Sci Rep, 6, 2016
|
|
7UBF
 
 | |
7UBD
 
 | |
7UBI
 
 | |
7UZL
 
 | |
7UBC
 
 | |
7UBE
 
 | |
7UBG
 
 | |
7UBH
 
 | |
2N73
 
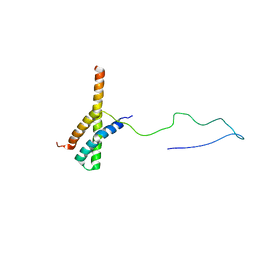 | | Solution structure of the ACBD3:PI4KB complex | | Descriptor: | Golgi resident protein GCP60, Phosphatidylinositol 4-kinase beta | | Authors: | Veverka, V, Hexnerova, R. | | Deposit date: | 2015-09-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights and in vitro reconstitution of membrane targeting and activation of human PI4KB by the ACBD3 protein.
Sci Rep, 6, 2016
|
|
8AXH
 
 | | Crystal structure of a MUC1-like glycopeptide containing the unnatural L-4-hydroxynorvaline in complex with scFv-SM3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, Mucin-1 subunit alpha, ... | | Authors: | Bermejo, I, Corzana, F, Hurtado-Guerrero, R. | | Deposit date: | 2022-08-31 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Approach for the Development of MUC1-Glycopeptide-Based Cancer Vaccines with Predictable Responses.
Jacs Au, 4, 2024
|
|
6R5M
 
 | | Crystal structure of toxin MT9 from mamba venom | | Descriptor: | ACETYL GROUP, Dendroaspis polylepis MT9, GLYCEROL, ... | | Authors: | Stura, E.A, Tepshi, L, Ciolek, J, Triquigneaux, M, Zoukimian, C, De Waard, M, Beroud, R, Servent, D, Gilles, N, Legrand, P, Ciccone, L. | | Deposit date: | 2019-03-25 | | Release date: | 2020-02-12 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MT9, a natural peptide from black mamba venom antagonizes the muscarinic type 2 receptor and reverses the M2R-agonist-induced relaxation in rat and human arteries
Biomed Pharmacother, 150, 2022
|
|
4MVF
 
 | | Crystal Structure of Plasmodium falciparum CDPK2 complexed with inhibitor staurosporine | | Descriptor: | CALCIUM ION, Calcium-dependent protein kinase 2, GLYCEROL, ... | | Authors: | Lauciello, L, Pernot, L, Scapozza, L, Perozzo, R. | | Deposit date: | 2013-09-24 | | Release date: | 2014-09-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | P. falciparum Calcium-Dependent Protein Kinase 2 (PfCDPK2): First Crystal Structure and Virtual Ligand Screening
To be Published
|
|
4OQL
 
 | | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with dF-EdU | | Descriptor: | 2'-deoxy-5-ethynyl-2',2'-difluorouridine, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Neef, A.B, Westermaier, Y, Perozzo, R, Luedtke, N, Scapozza, L. | | Deposit date: | 2014-02-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with dF-EdU
To be Published
|
|
8P0O
 
 | | Crystal structure of AaNGT complexed to UDP-Gal | | Descriptor: | 1,2-ETHANEDIOL, Adhesin, GALACTOSE-URIDINE-5'-DIPHOSPHATE | | Authors: | Pinello, B, Macias-Leon, J, Rovira, C, Hurtado-Guerrero, R. | | Deposit date: | 2023-05-10 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular basis for bacterial N-glycosylation by a soluble HMW1C-like N-glycosyltransferase.
Nat Commun, 14, 2023
|
|
8P0Q
 
 | | Crystal structure of AaNGT complexed to UDP and a peptide | | Descriptor: | Adhesin, PHE-GLY-ASN-TRP-THR-THR, URIDINE-5'-DIPHOSPHATE | | Authors: | Piniello, B, Macias-Leon, J, Rovira, C, Hurtado-Guerrero, R. | | Deposit date: | 2023-05-10 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for bacterial N-glycosylation by a soluble HMW1C-like N-glycosyltransferase.
Nat Commun, 14, 2023
|
|
8AY1
 
 | | Crystal structure of the C. elegans POFUT2 (CePoFUT2) triple mutant (R298K-R299K-A418C) in complex with the Rattus norvegicus TSR4 single mutant (E10C) from F-spondin | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 2,Spondin-1, ... | | Authors: | Hurtado-Guerrero, R, Merino, P. | | Deposit date: | 2022-09-01 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The Essential Role of Water Molecules in the Reaction Mechanism of Protein O-Fucosyltransferase 2.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8P0P
 
 | | Crystal structure of AaNGT complexed to UDP-2F-Glucose | | Descriptor: | Adhesin, URIDINE-5'-DIPHOSPHATE, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | Authors: | Piniello, B, Macias-Leon, J, Rovira, C, Hurtado-Guerrero, R. | | Deposit date: | 2023-05-10 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Molecular basis for bacterial N-glycosylation by a soluble HMW1C-like N-glycosyltransferase.
Nat Commun, 14, 2023
|
|
8PE1
 
 | | Crystal structure of Gel4 in complex with Nanobody 4 | | Descriptor: | 1,3-beta-glucanosyltransferase, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody 4, ... | | Authors: | Macias-Leon, J, Redrado-Hernandez, S, Castro-Lopez, J, Sanz, A.B, Arias, M, Farkas, V, Vincke, C, Muyldermans, S, Pardo, J, Arroyo, J, Galvez, E, Hurtado-Guerrero, R. | | Deposit date: | 2023-06-13 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Broad Protection against Invasive Fungal Disease from a Nanobody Targeting the Active Site of Fungal beta-1,3-Glucanosyltransferases.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
4OQM
 
 | | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with F-ARA-EdU | | Descriptor: | 1-(2-deoxy-2-fluoro-beta-D-arabinofuranosyl)-5-ethynylpyrimidine-2,4(1H,3H)-dione, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Neef, A.B, Westermaier, Y, Perozzo, R, Luedtke, N, Scapozza, L. | | Deposit date: | 2014-02-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with F-ARA-EdU
To be Published
|
|
8PE2
 
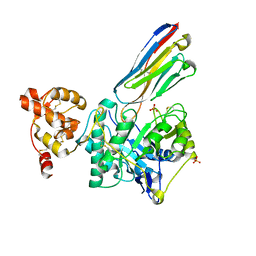 | | Crystal structure of Gel4 in complex with Nanobody 3 | | Descriptor: | 1,3-beta-glucanosyltransferase, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody 3, ... | | Authors: | Macias-Leon, J, Redrado-Hernandez, S, Castro-Lopez, J, Sanz, A.B, Arias, M, Farkas, V, Vincke, C, Muyldermans, S, Pardo, J, Arroyo, J, Galvez, E, Hurtado-Guerrero, R. | | Deposit date: | 2023-06-13 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Broad Protection against Invasive Fungal Disease from a Nanobody Targeting the Active Site of Fungal beta-1,3-Glucanosyltransferases.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6GW7
 
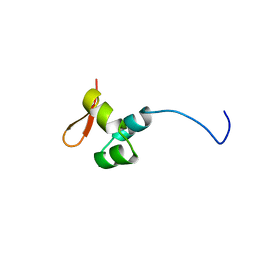 | | The CTD of HpDprA, a DNA binding Winged Helix domain which do not bind dsDNA | | Descriptor: | DNA protecting protein DprA | | Authors: | Lisboa, J, Celma, L, Sanchez, D, Marquis, M, Andreani, J, Guerois, R, Ochsenbein, F, Durand, D, Marsin, S, Cuniasse, P, Radicella, J.P, Quevillon-Cheruel, S. | | Deposit date: | 2018-06-22 | | Release date: | 2019-04-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The C-terminal domain of HpDprA is a DNA-binding winged helix domain that does not bind double-stranded DNA.
Febs J., 286, 2019
|
|
4NFU
 
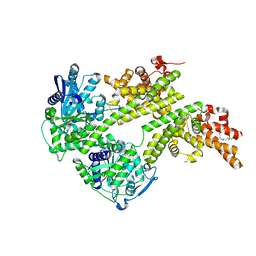 | | Structure of the central plant immunity signaling node EDS1 in complex with its interaction partner SAG101 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, EDS1, ISOPROPYL ALCOHOL, ... | | Authors: | Wagner, S, Stuttmann, J, Rietz, S, Guerois, R, Niefind, K, Parker, J.E. | | Deposit date: | 2013-11-01 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis for Signaling by Exclusive EDS1 Heteromeric Complexes with SAG101 or PAD4 in Plant Innate Immunity.
Cell Host Microbe, 14, 2013
|
|
4OQN
 
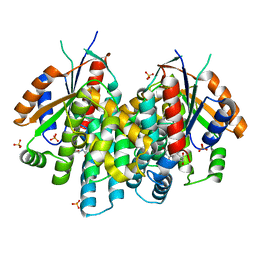 | | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with EdU | | Descriptor: | 2'-deoxy-5-ethynyluridine, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Neef, A.B, Westermaier, Y, Perozzo, R, Luedtke, N.W, Scapozza, L. | | Deposit date: | 2014-02-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with EdU
To be Published
|
|
