2NTI
 
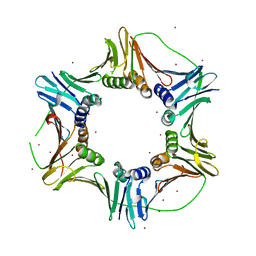 | | Crystal structure of PCNA123 heterotrimer. | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, BROMIDE ION, DNA polymerase sliding clamp A, ... | | Authors: | Hlinkova, V, Ling, H. | | Deposit date: | 2006-11-07 | | Release date: | 2007-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of monomeric, dimeric and trimeric PCNA: PCNA-ring assembly and opening.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1FD4
 
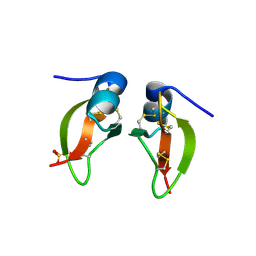 | | HUMAN BETA-DEFENSIN 2 | | Descriptor: | BETA-DEFENSIN 2, SULFATE ION | | Authors: | Hoover, D.M, Lubkowski, J. | | Deposit date: | 2000-07-19 | | Release date: | 2000-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of human beta-defensin-2 shows evidence of higher order oligomerization.
J.Biol.Chem., 275, 2000
|
|
1FD3
 
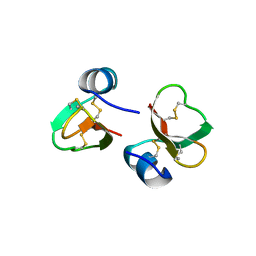 | | HUMAN BETA-DEFENSIN 2 | | Descriptor: | BETA-DEFENSIN 2, SULFATE ION | | Authors: | Hoover, D.M, Lubkowski, J. | | Deposit date: | 2000-07-19 | | Release date: | 2000-11-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structure of human beta-defensin-2 shows evidence of higher order oligomerization.
J.Biol.Chem., 275, 2000
|
|
5UI8
 
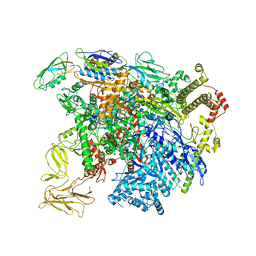 | | structure of sigmaN-holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darst, S.A, Campbell, E.A. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.76 Å) | | Cite: | Crystal structure of Aquifex aeolicus sigma (N) bound to promoter DNA and the structure of sigma (N)-holoenzyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6WWX
 
 | |
6WV2
 
 | |
6WXA
 
 | | Crystal structure of truncated Streptococcal bacteriophage hyaluronidase complexed with unsaturated hyaluronan hexa-saccharides | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Deivanayagam, C, Schormann, N. | | Deposit date: | 2020-05-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Streptococcal Bacteriophage Hyaluronidase:
Presence of a Prokaryotic Collagen and Elucidation of Catalytic Mechanism
To Be Published
|
|
6X3M
 
 | | Crystal structure of full-length Streptococcal bacteriophage hyaluronidase in complex with unsaturated hyaluronan octa-saccharides | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hyaluronoglucosaminidase | | Authors: | Deivanayagam, C, Schormann, N. | | Deposit date: | 2020-05-21 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.581 Å) | | Cite: | Crystal Structure of Streptococcal Bacteriophage Hyaluronidase:
Presence of a Prokaryotic Collagen and Elucidation of Catalytic Mechanism
To Be Published
|
|
3T51
 
 | | Crystal structures of the pre-extrusion and extrusion states of the CusBA adaptor-transporter complex | | Descriptor: | COPPER (II) ION, Cation efflux system protein CusA, Cation efflux system protein CusB | | Authors: | Su, C.-C, Long, F, Yu, E.W. | | Deposit date: | 2011-07-26 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Charged Amino Acids (R83, E567, D617, E625, R669, and K678) of CusA Are Required for Metal Ion Transport in the Cus Efflux System.
J.Mol.Biol., 422, 2012
|
|
3V78
 
 | | Crystal Structure of Transcriptional Regulator | | Descriptor: | ETHIDIUM, PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY DEOR-FAMILY) | | Authors: | Do, S.V, Bolla, J.R, Chen, X, Yu, E.W. | | Deposit date: | 2011-12-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural and functional analysis of the transcriptional regulator Rv3066 of Mycobacterium tuberculosis.
Nucleic Acids Res., 40, 2012
|
|
3V6G
 
 | | Crystal Structure of Transcriptional Regulator | | Descriptor: | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY DEOR-FAMILY) | | Authors: | Do, S.V, Bolla, J.R, Chen, X, Yu, E.W. | | Deposit date: | 2011-12-19 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Structural and functional analysis of the transcriptional regulator Rv3066 of Mycobacterium tuberculosis.
Nucleic Acids Res., 40, 2012
|
|
3T53
 
 | | Crystal structures of the extrusion state of the CusBA adaptor-transporter complex | | Descriptor: | COPPER (II) ION, Cation efflux system protein CusA, Cation efflux system protein CusB | | Authors: | Su, C.-C, Long, F, Yu, E.W. | | Deposit date: | 2011-07-26 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Charged Amino Acids (R83, E567, D617, E625, R669, and K678) of CusA Are Required for Metal Ion Transport in the Cus Efflux System.
J.Mol.Biol., 422, 2012
|
|
3T56
 
 | | Crystal structure of the pre-extrusion state of the CusBA adaptor-transporter complex | | Descriptor: | COPPER (II) ION, Cation efflux system protein CusA, Cation efflux system protein CusB | | Authors: | Su, C.-C, Long, F, Yu, E.W. | | Deposit date: | 2011-07-26 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Charged Amino Acids (R83, E567, D617, E625, R669, and K678) of CusA Are Required for Metal Ion Transport in the Cus Efflux System.
J.Mol.Biol., 422, 2012
|
|
5BSA
 
 | | Structure of histone H3/H4 in complex with Spt2 | | Descriptor: | Histone H3.2, Histone H4, Protein SPT2 homolog | | Authors: | Chen, S, Patel, D.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.611 Å) | | Cite: | Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2.
Genes Dev., 29, 2015
|
|
5BS7
 
 | | Structure of histone H3/H4 in complex with Spt2 | | Descriptor: | Histone H3.2, Histone H4, Protein SPT2 homolog, ... | | Authors: | Chen, S, Patel, D.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2.
Genes Dev., 29, 2015
|
|
4JYO
 
 | | Structural basis for angiopoietin-1 mediated signaling initiation | | Descriptor: | Angiopoietin-1, CALCIUM ION | | Authors: | Yu, X, Seegar, T.C.M, Dalton, A.C, Tzvetkova-Robev, D, Goldgur, Y, Nikolov, D.B, Barton, W.A. | | Deposit date: | 2013-03-31 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for angiopoietin-1-mediated signaling initiation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JZC
 
 | | Angiopoietin-2 fibrinogen domain TAG mutant | | Descriptor: | Angiopoietin-2 | | Authors: | Yu, X, Seegar, T.C.M, Dalton, A.C, Tzvetkova-Robev, D, Goldgur, Y, Nikolov, D.B, Barton, W.A. | | Deposit date: | 2013-04-02 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for angiopoietin-1-mediated signaling initiation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K0V
 
 | | Structural basis for angiopoietin-1 mediated signaling initiation | | Descriptor: | Angiopoietin-1, TEK tyrosine kinase variant | | Authors: | Yu, X, Seegar, T.C.M, Dalton, A.C, Tzvetkova-Robev, D, Goldgur, Y, Nikolov, D.B, Barton, W.A. | | Deposit date: | 2013-04-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.51 Å) | | Cite: | Structural basis for angiopoietin-1-mediated signaling initiation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5KHS
 
 | |
5KHN
 
 | |
4NB5
 
 | |
4MT1
 
 | |
4NN1
 
 | |
5FG1
 
 | |
5LQ3
 
 | |
