2W1L
 
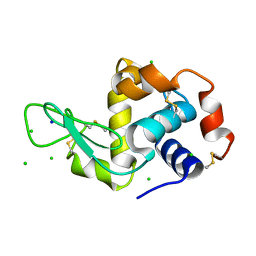 | | THE INTERDEPENDENCE OF WAVELENGTH, REDUNDANCY AND DOSE IN SULFUR SAD EXPERIMENTS: 0.979 a wavelength 991 images data | | Descriptor: | CHLORIDE ION, LYSOZYME C, SODIUM ION | | Authors: | Cianci, M, Helliwell, J.R, Suzuki, A. | | Deposit date: | 2008-10-17 | | Release date: | 2008-10-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The Interdependence of Wavelength, Redundancy and Dose in Sulfur Sad Experiments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2W2G
 
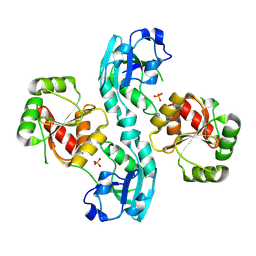 | | Human SARS coronavirus unique domain | | Descriptor: | NON-STRUCTURAL PROTEIN 3, SULFATE ION | | Authors: | Tan, J, Vonrhein, C, Smart, O.S, Bricogne, G, Bollati, M, Kusov, Y, Hansen, G, Mesters, J.R, Schmidt, C.L, Hilgenfeld, R. | | Deposit date: | 2008-10-30 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The Sars-Unique Domain (Sud) of Sars Coronavirus Contains Two Macrodomains that Bind G-Quadruplexes.
Plos Pathog., 5, 2009
|
|
2W1X
 
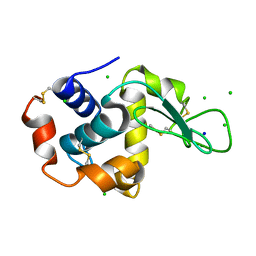 | | The interdependence of wavelength, redundancy and dose in sulfur SAD experiments: 1.284 A wavelength 360 images data | | Descriptor: | CHLORIDE ION, LYSOZYME C, SODIUM ION | | Authors: | Cianci, M, Helliwell, J.R, Suzuki, A. | | Deposit date: | 2008-10-21 | | Release date: | 2008-11-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Interdependence of Wavelength, Redundancy and Dose in Sulfur Sad Experiments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2W6Z
 
 | |
2W6O
 
 | | Crystal structure of Biotin carboxylase from E. coli in complex with 4-Amino-7,7-dimethyl-7,8-dihydro-quinazolinone fragment | | Descriptor: | 4-amino-7,7-dimethyl-7,8-dihydroquinazolin-5(6H)-one, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
2W5Z
 
 | | Ternary Complex of the Mixed Lineage Leukaemia (MLL1) SET Domain with the cofactor product S-Adenosylhomocysteine and histone peptide. | | Descriptor: | GLYCEROL, HISTONE PEPTIDE, HISTONE-LYSINE N-METHYLTRANSFERASE HRX, ... | | Authors: | Southall, S.M, Wong, P.S, Odho, Z, Roe, S.M, Wilson, J.R. | | Deposit date: | 2008-12-15 | | Release date: | 2009-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Requirement of Additional Factors for Mll1 Set Domain Activity and Recognition of Epigenetic Marks.
Mol.Cell, 33, 2009
|
|
2W9S
 
 | | Staphylococcus aureus S1:DHFR in complex with trimethoprim | | Descriptor: | DIHYDROFOLATE REDUCTASE TYPE 1 FROM TN4003, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Soutter, H.H, Miller, J.R. | | Deposit date: | 2009-01-28 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Comparison of Chromosomal and Exogenous Dihydrofolate Reductase from Staphylococcus Aureus in Complex with the Potent Inhibitor Trimethoprim.
Proteins, 76, 2009
|
|
2VRI
 
 | |
2W6P
 
 | |
2WCT
 
 | | human SARS coronavirus unique domain (triclinic form) | | Descriptor: | NON-STRUCTURAL PROTEIN 3 | | Authors: | Tan, J, Vonrhein, C, Smart, O.S, Bricogne, G, Bollati, M, Hansen, G, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2009-03-16 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The Sars-Unique Domain (Sud) of Sars Coronavirus Contains Two Macrodomains that Bind G-Quadruplexes.
Plos Pathog., 5, 2009
|
|
2W5Y
 
 | | Binary Complex of the Mixed Lineage Leukaemia (MLL1) SET Domain with the cofactor product S-Adenosylhomocysteine. | | Descriptor: | HISTONE-LYSINE N-METHYLTRANSFERASE HRX, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Southall, S.M, Wong, P.S, Odho, Z, Roe, S.M, WIlson, J.R. | | Deposit date: | 2008-12-15 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Requirement of Additional Factors for Mll1 Set Domain Activity and Recognition of Epigenetic Marks.
Mol.Cell, 33, 2009
|
|
2W6M
 
 | |
1AKX
 
 | |
1AJU
 
 | |
5M10
 
 | |
6ZUJ
 
 | | Human serine racemase holoenzyme from 20% DMSO soak (XChem crystallographic fragment screen). | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Koulouris, C.R, Bax, B.D, Roe, S.M, Atack, J.R. | | Deposit date: | 2020-07-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
6ZSP
 
 | | Human serine racemase bound to ATP and malonate. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Koulouris, C.R, Bax, B.D, Roe, S.M, Atack, J.R. | | Deposit date: | 2020-07-16 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
8BCQ
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (native crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION | | Authors: | Benas, P, Jaramillo Ponce, J.R, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
8BHD
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (Tbxo4 derivative crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION, ... | | Authors: | Benas, P, Jaramillo Ponce, J.R, Legrand, P, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
7F9O
 
 | | PSI-NDH supercomplex of Barley | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2021-07-04 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
7EW6
 
 | | Barley photosystem I-LHCI-Lhca5 supercomplex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2021-05-24 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
5OV3
 
 | | Structure of the RbBP5 beta-propeller domain | | Descriptor: | Retinoblastoma-binding protein 5, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Mittal, A, Zhang, Y, Gamblin, S.J, Wilson, J.R. | | Deposit date: | 2017-08-27 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of the RbBP5 beta-propeller domain reveals a surface with potential nucleic acid binding sites.
Nucleic Acids Res., 46, 2018
|
|
7W2X
 
 | | Crystal structure of TxGH116 R786K mutant from Thermoanaerobacterium xylanolyticum | | Descriptor: | CALCIUM ION, GLYCEROL, Glucosylceramidase, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
7W2S
 
 | | Crystal structure of TxGH116 E730A mutant from Thermoanaerobacterium xylanolyticum with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
7W2W
 
 | | Crystal structure of TxGH116 R786K mutant from Thermoanaerobacterium xylanolyticum with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
