8C4A
 
 | |
6RKF
 
 | | Structure of human DASPO | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-aspartate oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chaves-Sanjuan, A, Nardini, M. | | Deposit date: | 2019-04-30 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.219 Å) | | Cite: | Structure and kinetic properties of human d-aspartate oxidase, the enzyme-controlling d-aspartate levels in brain.
Faseb J., 34, 2020
|
|
2MZ2
 
 | |
2MS5
 
 | | Structural dynamics of double-helical RNA having CAG motif | | Descriptor: | RNA_(5'-R(P*CP*CP*GP*CP*AP*GP*CP*GP*G)-3') | | Authors: | Kumar, A, Tawani, A. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insights Reveal the Dynamics of the Repeating r(CAG) Transcript Found in Huntington's Disease (HD) and Spinocerebellar Ataxias (SCAs)
Plos One, 10, 2015
|
|
8BXT
 
 | | Structure of StayGold | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, StayGold | | Authors: | Ivorra-Molla, E, Akhuli, D, Crow, A. | | Deposit date: | 2022-12-09 | | Release date: | 2023-07-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A monomeric StayGold fluorescent protein.
Nat.Biotechnol., 2023
|
|
1IE4
 
 | | RAT TRANSTHYRETIN COMPLEX WITH THYROXINE (T4) | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, TRANSTHYRETIN | | Authors: | Wojtczak, A. | | Deposit date: | 2001-04-06 | | Release date: | 2002-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of rat transthyretin (rTTR) complex with thyroxine at 2.5 A resolution: first non-biased insight into thyroxine binding reveals different hormone orientation in two binding sites.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6RH7
 
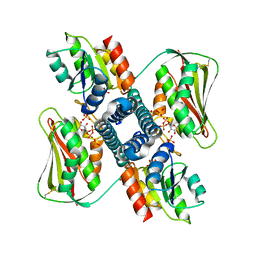 | | Revisiting pH-gated conformational switch. Complex HK853 mutant H260A -RR468 mutant D53A pH 7.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
3VCX
 
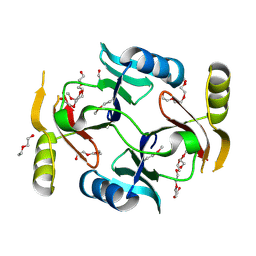 | | Crystal structure of a putative glyoxalase/bleomycin resistance protein from Rhodopseudomonas palustris CGA009 | | Descriptor: | Glyoxalase/Bleomycin resistance protein/dioxygenase domain, TETRAETHYLENE GLYCOL | | Authors: | Stogios, P.J, Chang, C, Evdokimova, E, Egorova, O, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-04 | | Release date: | 2012-01-18 | | Last modified: | 2012-01-25 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structure of a putative glyoxalase/bleomycin resistance protein from Rhodopseudomonas palustris CGA009
To be Published
|
|
6RH1
 
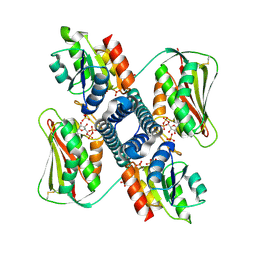 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 D53A pH 7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
2MXH
 
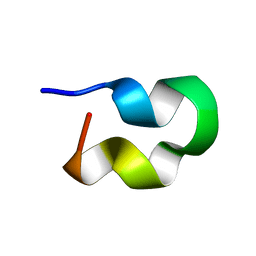 | |
1WS5
 
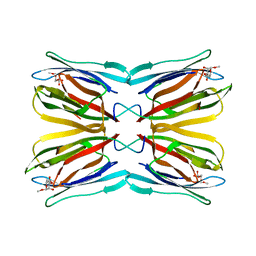 | | Crystal structure of Jacalin-Me-alpha-Mannose complex: Promiscuity vs Specificity | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-3 chain, methyl alpha-D-mannopyranoside | | Authors: | Jeyaprakash, A.A, Jayashree, G, Mahanta, S.K, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2004-10-31 | | Release date: | 2005-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the energetics of jacalin-sugar interactions: promiscuity versus specificity
J.Mol.Biol., 347, 2005
|
|
2DRM
 
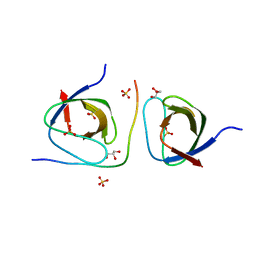 | |
3II7
 
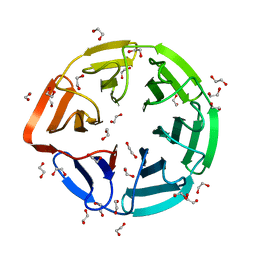 | | Crystal structure of the kelch domain of human KLHL7 | | Descriptor: | 1,2-ETHANEDIOL, Kelch-like protein 7 | | Authors: | Chaikuad, A, Thangaratnarajah, C, Cooper, C.D.O, Ugochukwu, E, Muniz, J.R.C, Krojer, T, Sethi, R, Pike, A.C.W, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-31 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases.
J.Biol.Chem., 288, 2013
|
|
3QXH
 
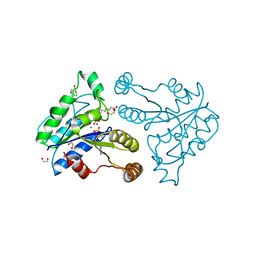 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with ADP and 8-aminocaprylic acid | | Descriptor: | 1,2-ETHANEDIOL, 8-aminooctanoic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Minor, C, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QVI
 
 | | Crystal structure of KNI-10395 bound histo-aspartic protease (HAP) from Plasmodium falciparum | | Descriptor: | (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-{[S-methyl-N-(phenylacetyl)-L-cysteinyl]amino}-4-phenylbutanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-02-25 | | Release date: | 2011-10-12 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the activation and inhibition of histo-aspartic protease from Plasmodium falciparum.
Biochemistry, 50, 2011
|
|
3QX5
 
 | | Thrombin Inhibition by Pyridin Derivatives | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(4-chloro-1-methylpyridinium-2-yl)methyl]-L-prolinamide, GLYCEROL, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-03-01 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Impact of ligand and protein desolvation on ligand binding to the S1 pocket of thrombin
J.Mol.Biol., 418, 2012
|
|
3QXS
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with ANP | | Descriptor: | 1,2-ETHANEDIOL, Dethiobiotin synthetase, MAGNESIUM ION, ... | | Authors: | Klimecka, M.M, Porebski, P.J, Chruszcz, M, Jablonska, K, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QQZ
 
 | | Crystal structure of the C-terminal domain of the yjiK protein from Escherichia coli CFT073 | | Descriptor: | CALCIUM ION, Putative uncharacterized protein yjiK | | Authors: | Stein, A, Chhor, G, Nocek, B, Fenske, R.J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-16 | | Release date: | 2011-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the C-terminal domain of the yjiK protein from Escherichia coli CFT073
TO BE PUBLISHED
|
|
3VJJ
 
 | | Crystal Structure Analysis of the P9-1 | | Descriptor: | P9-1 | | Authors: | Akita, F, Higashiura, A, Suzuki, M, Tsukihara, T, Nakagawa, A, Omura, T. | | Deposit date: | 2011-10-24 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analysis reveals octamerization of viroplasm matrix protein P9-1 of Rice black streaked dwarf virus
J.Virol., 86, 2012
|
|
8ERN
 
 | | Cyclin-free CDK2 in complex with Cpd21 | | Descriptor: | Cyclin-dependent kinase 2, GLYCEROL, N,3-dimethyl-4-({7-[(2S)-2-methylpyrrolidine-1-carbonyl]quinazolin-2-yl}amino)benzene-1-sulfonamide | | Authors: | Murray, J.M, Oh, A. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Cyclin-free CDK2 in complex with Cpd21
To Be Published
|
|
1INV
 
 | | A SIALIC ACID DERIVED PHOSPHONATE ANALOG INHIBITS DIFFERENT STRAINS OF INFLUENZA VIRUS NEURAMINIDASE WITH DIFFERENT EFFICIENCIES | | Descriptor: | (1R)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | White, C.L, Janakiraman, M.N, Laver, W.G, Philippon, C, Vasella, A, Air, G.M, Luo, M. | | Deposit date: | 1994-09-26 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A sialic acid-derived phosphonate analog inhibits different strains of influenza virus neuraminidase with different efficiencies.
J.Mol.Biol., 245, 1995
|
|
2MPF
 
 | | Solution structure human HCN2 CNBD in the cAMP-unbound state | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Saponaro, A, Pauleta, S.R, Cantini, F, Matzapetakis, M, Hammann, C, Banci, L, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2014-05-16 | | Release date: | 2014-09-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the mutual antagonism of cAMP and TRIP8b in regulating HCN channel function.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6S1N
 
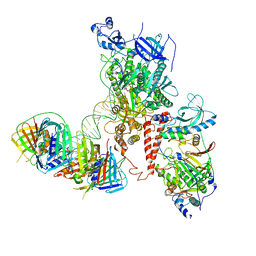 | | Human polymerase delta holoenzyme Conformer 2 | | Descriptor: | DNA polymerase delta catalytic subunit, DNA polymerase delta subunit 2, DNA polymerase delta subunit 3, ... | | Authors: | Lancey, C, Hamdan, S.M, De Biasio, A. | | Deposit date: | 2019-06-19 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.86 Å) | | Cite: | Structure of the processive human Pol delta holoenzyme.
Nat Commun, 11, 2020
|
|
3AGW
 
 | |
6S2V
 
 | |
