6QX4
 
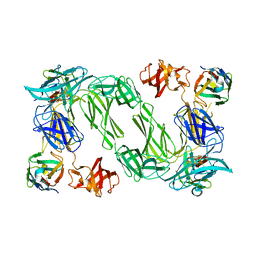 | |
5SGG
 
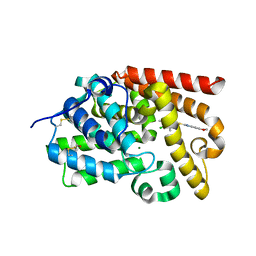 | | Crystal Structure of human phosphodiesterase 10 in complex with 2-[(4-chloro-3,5-dimethylpyridin-2-yl)methylsulfanyl]-6,7-dihydro-3H-furo[3,2-f]benzimidazole | | Descriptor: | 2-{[(4-chloro-3,5-dimethylpyridin-2-yl)methyl]sulfanyl}-6,7-dihydro-1H-furo[2,3-f]benzimidazole, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Krasso, A, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SH4
 
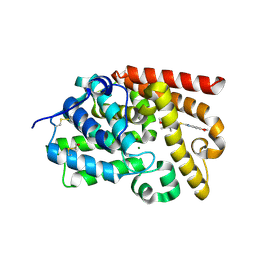 | | Crystal Structure of human phosphodiesterase 10 in complex with 6-[(5-methoxy-3,4,6-trimethylpyridin-2-yl)methylsulfanyl]-5H-[1,3]dioxolo[4,5-f]benzimidazole | | Descriptor: | 6-{[(5-methoxy-3,4,6-trimethylpyridin-2-yl)methyl]sulfanyl}-2H,5H-[1,3]dioxolo[4,5-f]benzimidazole, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Krasso, A, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
2MKO
 
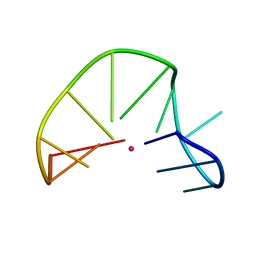 | | G-triplex structure and formation propensity | | Descriptor: | DNA_(5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*G)-3'), POTASSIUM ION | | Authors: | Cerofolini, L, Fragai, M, Giachetti, A, Limongelli, V, Luchinat, C, Novellino, E, Parrinello, M, Randazzo, A. | | Deposit date: | 2014-02-11 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | G-triplex structure and formation propensity.
Nucleic Acids Res., 42, 2014
|
|
3QRV
 
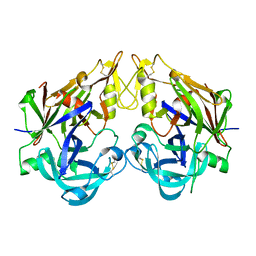 | |
4FVO
 
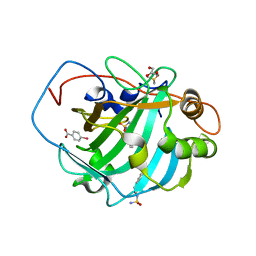 | |
3QTO
 
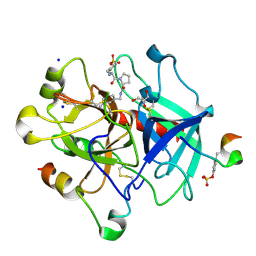 | | Thrombin Inhibition by Pyridin Derivatives | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(1-methylpyridinium-3-yl)methyl]-L-prolinamide, GLYCEROL, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-02-23 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Impact of ligand and protein desolvation on ligand binding to the S1 pocket of thrombin
J.Mol.Biol., 418, 2012
|
|
8ET3
 
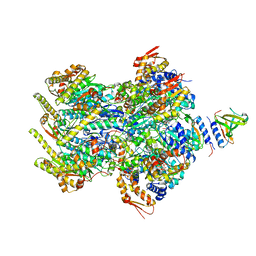 | | Cryo-EM structure of a delivery complex containing the SspB adaptor, an ssrA-tagged substrate, and the AAA+ ClpXP protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Ghanbarpour, A, Fei, X, Davis, J.H, Sauer, R.T. | | Deposit date: | 2022-10-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The SspB adaptor drives structural changes in the AAA+ ClpXP protease during ssrA-tagged substrate delivery.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3QVC
 
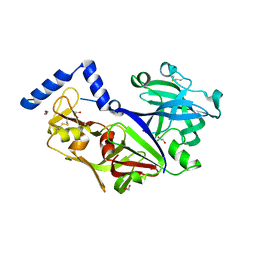 | |
3QVQ
 
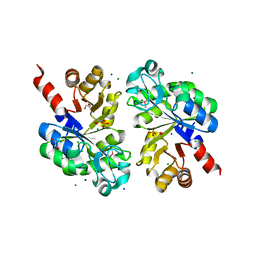 | | The structure of an Oleispira antarctica phosphodiesterase OLEI02445 in complex with the product sn-glycerol-3-phosphate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Kagan, O, Evdokimova, E, Cuff, M.E, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-25 | | Release date: | 2011-04-13 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | The structure of an Oleispira antarctica phosphodiesterase OLEI02445 in complex with the product sn-glycerol-3-phosphate
To be Published
|
|
8EEQ
 
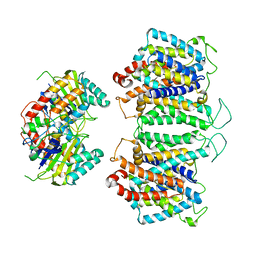 | | CryoEM structures of bAE1 captured in multiple states. | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-09-07 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
3Q5J
 
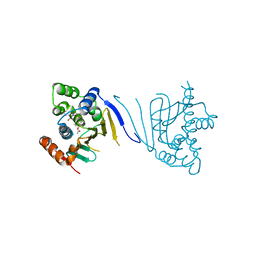 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of 17-DMAP-geldanamycin | | Descriptor: | (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-19-{[3-(dimethylamino)propyl]amino}-13-hydroxy-8,14-dimethoxy-4,10,12,16-tetramethyl-3,20,22-trioxo-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate, Heat shock protein 83-1 | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-28 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of 17-DMAP-geldanamycin.
To be Published
|
|
2CXU
 
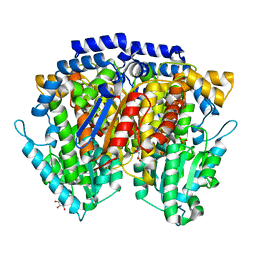 | | Crystal structure of mouse AMF / M6P complex | | Descriptor: | GLYCEROL, Glucose-6-phosphate isomerase, PHOSPHATE ION | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
8F0W
 
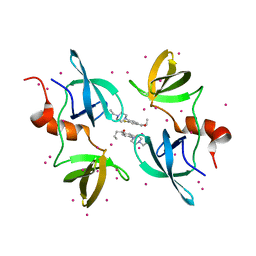 | | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956 | | Descriptor: | 1-[4-(4-ethylpiperazin-1-yl)-3-fluorophenyl]butan-1-one, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | The, J, Hong, Z, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-04 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956
to be published
|
|
3IP5
 
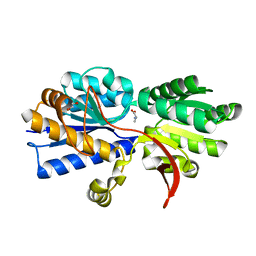 | | Structure of Atu2422-GABA receptor in complex with alanine | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), ALANINE, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
8F4A
 
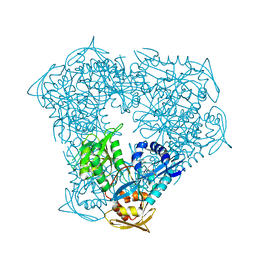 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with chlorhexidine | | Descriptor: | 1-[6-[azanylidene-[[azanylidene-[[(4-chlorophenyl)amino]methyl]-$l^{4}-azanyl]methyl]-$l^{4}-azanyl]hexyl]-3-[~{N}-(4-chlorophenyl)carbamimidoyl]guanidine, GLYCEROL, N-acetyltransferase Eis | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-10 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
3IP7
 
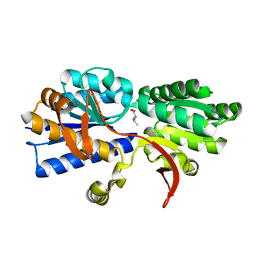 | | Structure of Atu2422-GABA receptor in complex with valine | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), CALCIUM ION, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
8F55
 
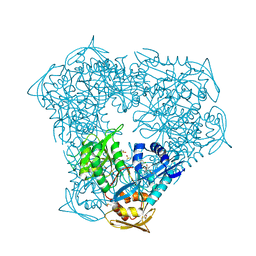 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis H37Rv in complex with inhibitor SGT1614 | | Descriptor: | (2E)-2-[(4-chlorophenyl)methylidene]hydrazine-1-carboximidamide, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
2CXT
 
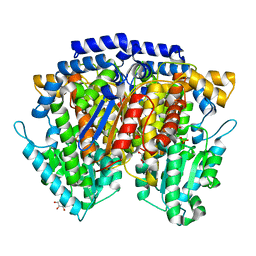 | | Crystal structure of mouse AMF / F6P complex | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, GLYCEROL, Glucose-6-phosphate isomerase | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
7M6M
 
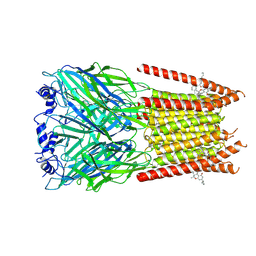 | | Full length alpha1 Glycine receptor in presence of 32uM Tetrahydrocannabinol | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1 | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
8F57
 
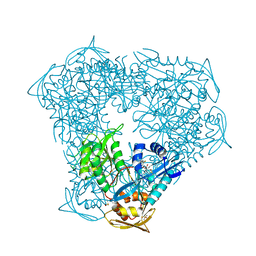 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with inhibitor SGT1615 | | Descriptor: | (1E)-1-[(4-chlorophenyl)methylidene]-2-(3-fluorophenyl)hydrazine, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-12 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
8EM7
 
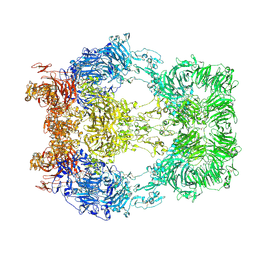 | | Cryo-EM structure of LRP2 at pH 5.2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Beenken, A, Cerutti, G, Fitzpatrick, A.W, Barasch, J, Shapiro, L. | | Deposit date: | 2022-09-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of LRP2 reveal a molecular machine for endocytosis.
Cell, 186, 2023
|
|
7M6N
 
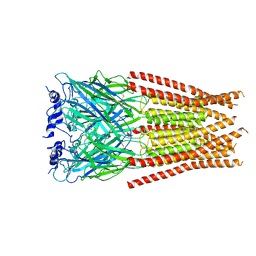 | |
2Z5F
 
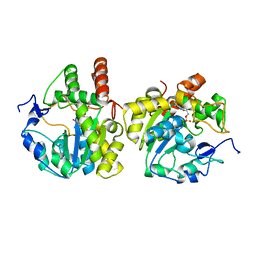 | | Human sulfotransferase Sult1b1 in complex with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase family cytosolic 1B member 1 | | Authors: | Dombrovski, L, Pegasova, T, Wu, H, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Sundstrom, M, Plotnikov, A.N, Bochkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-07 | | Release date: | 2007-08-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human sulfotransferases SULT1B1 and SULT1C1 complexed with the cofactor product adenosine-3'-5'-diphosphate (PAP)
Proteins, 64, 2006
|
|
7M6Q
 
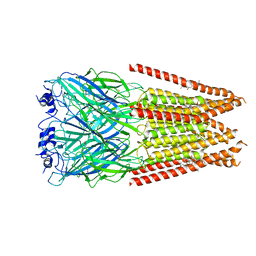 | | Full length alpha1 Glycine receptor in presence of 1mM Glycine and 32uM Tetrahydrocannabinol State 1 | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
