3ILR
 
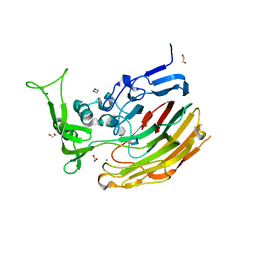 | | Structure of Heparinase I from Bacteroides thetaiotaomicron in complex with tetrasaccharide product | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(4-1)-4-deoxy-2-O-sulfo-beta-D-erythro-hex-4-enopyranuronic acid, 4-deoxy-2-O-sulfo-beta-D-erythro-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ... | | Authors: | Garron, M.L, Cygler, M, Shaya, D. | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural snapshots of heparin depolymerization by heparin lyase I.
J.Biol.Chem., 284, 2009
|
|
3ITD
 
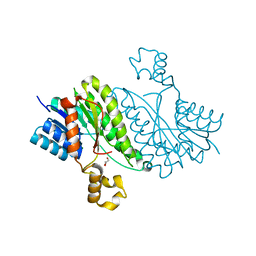 | |
3VTX
 
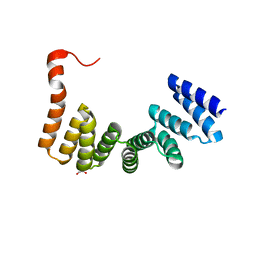 | | Crystal structure of MamA protein | | Descriptor: | GLYCEROL, MamA | | Authors: | Zeytuni, N, Baran, D, Davidov, G, Zarivach, R. | | Deposit date: | 2012-06-08 | | Release date: | 2012-10-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inter-phylum structural conservation of the magnetosome-associated TPR-containing protein, MamA
J.Struct.Biol., 180, 2012
|
|
1F0N
 
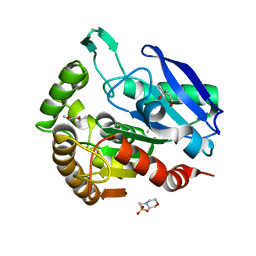 | | MYCOBACTERIUM TUBERCULOSIS ANTIGEN 85B | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ANTIGEN 85B | | Authors: | Anderson, D.H, Harth, G, Horwitz, M.A, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-05-16 | | Release date: | 2001-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An interfacial mechanism and a class of inhibitors inferred from two crystal structures of the Mycobacterium tuberculosis 30 kDa major secretory protein (Antigen 85B), a mycolyl transferase.
J.Mol.Biol., 307, 2001
|
|
3FGB
 
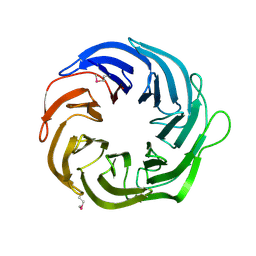 | | Crystal structure of the Q89ZH8_BACTN protein from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium target BtR289b. | | Descriptor: | uncharacterized protein Q89ZH8_BACTN | | Authors: | Vorobiev, S.M, Lew, S, Seetharaman, J, Lee, D, Foote, L.E, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-05 | | Release date: | 2008-12-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Q89ZH8_BACTN protein from Bacteroides thetaiotaomicron.
To be Published
|
|
1F0C
 
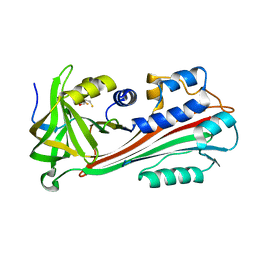 | | STRUCTURE OF THE VIRAL SERPIN CRMA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ICE INHIBITOR | | Authors: | Renatus, M, Zhou, Q, Stennicke, H.R, Snipas, S.J, Turk, D, Bankston, L.A, Liddington, R.C, Salvesen, G.S. | | Deposit date: | 2000-05-15 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of the apoptotic suppressor CrmA in its cleaved form.
Structure Fold.Des., 8, 2000
|
|
1F0E
 
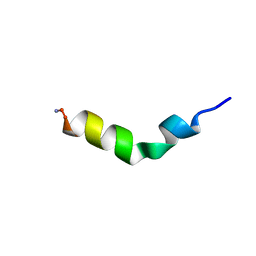 | | Cecropin A(1-8)-magainin 2(1-12) modified gig to P in dodecylphosphocholine micelles | | Descriptor: | CECROPIN A-MAGAININ 2 HYBRID PEPTIDE | | Authors: | Oh, D, Shin, S.Y, Lee, S, Kim, Y. | | Deposit date: | 2000-05-16 | | Release date: | 2000-06-14 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | Role of the hinge region and the tryptophan residue in the synthetic antimicrobial peptides, cecropin A(1-8)-magainin 2(1-12) and its analogues, on their antibiotic activities and structures.
Biochemistry, 39, 2000
|
|
1F0J
 
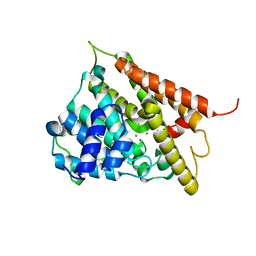 | | CATALYTIC DOMAIN OF HUMAN PHOSPHODIESTERASE 4B2B | | Descriptor: | ARSENIC, MAGNESIUM ION, PHOSPHODIESTERASE 4B, ... | | Authors: | Xu, R.X, Hassell, A.M, Vanderwall, D, Lambert, M.H, Holmes, W.D, Luther, M.A, Rocque, W.J, Milburn, M.V, Zhao, Y, Ke, H, Nolte, R.T. | | Deposit date: | 2000-05-16 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Atomic structure of PDE4: insights into phosphodiesterase mechanism and specificity.
Science, 288, 2000
|
|
1F0X
 
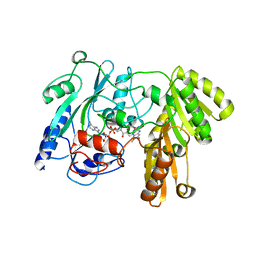 | | CRYSTAL STRUCTURE OF D-LACTATE DEHYDROGENASE, A PERIPHERAL MEMBRANE RESPIRATORY ENZYME. | | Descriptor: | D-LACTATE DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Dym, O, Pratt, E.A, Ho, C, Eisenberg, D. | | Deposit date: | 2000-05-17 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of D-lactate dehydrogenase, a peripheral membrane respiratory enzyme.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2MC9
 
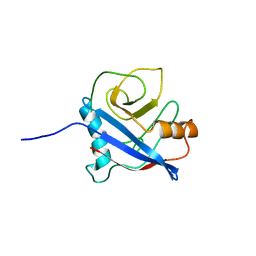 | | Cat r 1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Mueller, G, London, R, Ghosh, D. | | Deposit date: | 2013-08-16 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Primary identification, biochemical characterization, and immunologic properties of the allergenic pollen cyclophilin cat R 1.
J.Biol.Chem., 289, 2014
|
|
1F1D
 
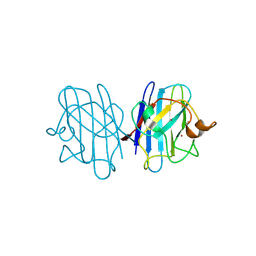 | | Crystal structure of yeast H46C cuznsod mutant | | Descriptor: | COPPER (II) ION, COPPER-ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Ogihara, N.L, Liu, H, Nersissian, A.M, Valentine, J.S, Eisenberg, D. | | Deposit date: | 2000-05-18 | | Release date: | 2002-12-18 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1F30
 
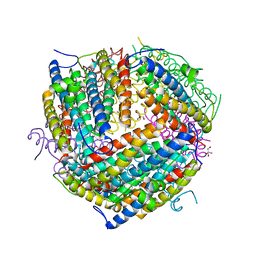 | | THE STRUCTURAL BASIS FOR DNA PROTECTION BY E. COLI DPS PROTEIN | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA PROTECTION DURING STARVATION PROTEIN, ZINC ION | | Authors: | Luo, J, Liu, D, White, M.A, Fox, R.O. | | Deposit date: | 2000-05-31 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Structural Basis for DNA Protection by E. coli Dps Protein
To be Published
|
|
3W0Q
 
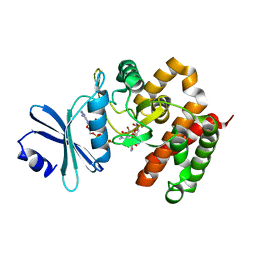 | | Crystal structure of a thermostable mutant of aminoglycoside phosphotransferase APH(4)-Ia (N203A), ternary complex with AMP-PNP and hygromycin B | | Descriptor: | HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
7ZJS
 
 | |
1F1T
 
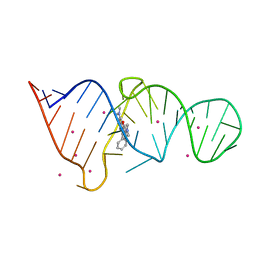 | | CRYSTAL STRUCTURE OF THE MALACHITE GREEN APTAMER COMPLEXED WITH TETRAMETHYL-ROSAMINE | | Descriptor: | MALACHITE GREEN APTAMER RNA, N,N'-TETRAMETHYL-ROSAMINE, STRONTIUM ION | | Authors: | Baugh, C, Grate, D, Wilson, C. | | Deposit date: | 2000-05-19 | | Release date: | 2000-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8 A crystal structure of the malachite green aptamer.
J.Mol.Biol., 301, 2000
|
|
6SGR
 
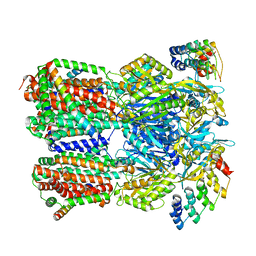 | | Cryo-EM structure of Escherichia coli AcrBZ and DARPin in Saposin A-nanodisc with cardiolipin | | Descriptor: | DARPin, Multidrug efflux pump accessory protein AcrZ, Multidrug efflux pump subunit AcrB | | Authors: | Szewczak-Harris, A, Du, D, Newman, C, Neuberger, A, Luisi, B.F. | | Deposit date: | 2019-08-05 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Interactions of a Bacterial RND Transporter with a Transmembrane Small Protein in a Lipid Environment.
Structure, 28, 2020
|
|
2M0T
 
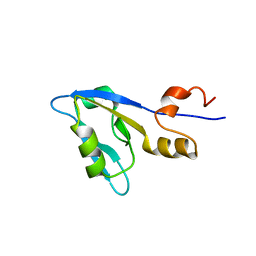 | |
6SBS
 
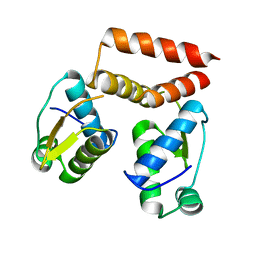 | | YtrA from Sulfolobus acidocaldarius, a GntR-family transcription factor | | Descriptor: | Regulatory protein | | Authors: | Lemmens, L, Valegard, K, Lindas, A.C, Peeters, E, Maes, D. | | Deposit date: | 2019-07-22 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | YtrASa, a GntR-Family Transcription Factor, Represses Two Genetic Loci Encoding Membrane Proteins inSulfolobus acidocaldarius.
Front Microbiol, 10, 2019
|
|
3W0P
 
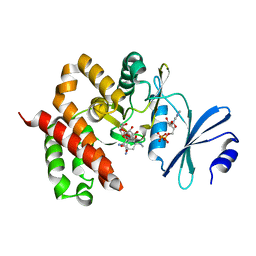 | | Crystal structure of a thermostable mutant of aminoglycoside phosphotransferase APH(4)-Ia (D198A), ternary complex with ADP and hygromycin B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
1F1H
 
 | |
6SHW
 
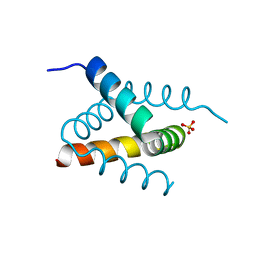 | | N-terminal domain of Drosophila X Virus VP3 | | Descriptor: | SULFATE ION, Structural polyprotein | | Authors: | Ferrero, D.S, Garriga, D, Guerra, P, Uson, I, Verdaguer, N. | | Deposit date: | 2019-08-08 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and dsRNA-binding activity of the Birnavirus Drosophila X Virus VP3 protein.
J.Virol., 2020
|
|
2M6U
 
 | | NMR Structure of CbpAN from Streptococcus pneumoniae | | Descriptor: | Choline binding protein A | | Authors: | Liu, A, Yan, H, Achila, D, Martinez-Hackert, E, Li, Y, Banerjee, R. | | Deposit date: | 2013-04-10 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of host specificity of complement Factor H recruitment by Streptococcus pneumoniae.
Biochem.J., 465, 2015
|
|
3IEC
 
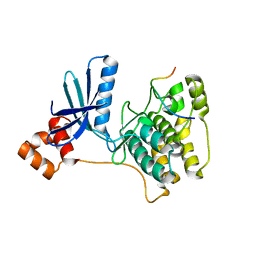 | |
2MLO
 
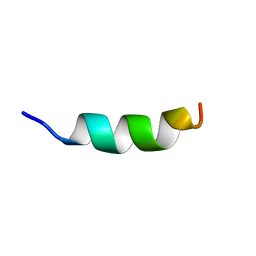 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a Membrane bound form | | Descriptor: | MCP-1 receptor | | Authors: | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | Deposit date: | 2014-03-04 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
7ZPN
 
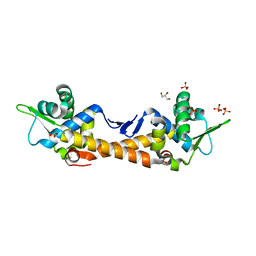 | | Crystal Structure of IscR from Dinoroseobacter shibae | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator, SULFATE ION | | Authors: | Lukat, P, Ploetzky, L, Blankenfeldt, W, Jahn, D, Haertig, E. | | Deposit date: | 2022-04-28 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Dinoroseobacter shibae IscR homolog acts as a repressor for iron acquisition genes
To Be Published
|
|
