7DTJ
 
 | |
3U9Q
 
 | |
4NIE
 
 | |
4LWV
 
 | | The 2.3A Crystal Structure of Humanized Xenopus MDM2 with RO5545353 | | Descriptor: | (2S,3R,4R,5R)-N-(4-carbamoyl-2-methoxyphenyl)-2'-chloro-4-(3-chloro-2-fluorophenyl)-2-(2,2-dimethylpropyl)-5'-oxo-4',5'-dihydrospiro[pyrrolidine-3,6'-thieno[3,2-b]pyrrole]-5-carboxamide, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Graves, B.J, Lukacs, C, Janson, C.A. | | Deposit date: | 2013-07-28 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Discovery of Potent and Orally Active p53-MDM2 Inhibitors RO5353 and RO2468 for Potential Clinical Development.
ACS MED.CHEM.LETT., 5, 2014
|
|
3UA0
 
 | | N-Terminal Domain of Bombyx mori Fibroin Mediates the Assembly of Silk in Response to pH Decrease | | Descriptor: | Fibroin heavy chain | | Authors: | He, Y.-X, Zhang, N.-N, Chen, B.-Y, Li, W.-F, Chen, Y.-X, Zhou, C.-Z. | | Deposit date: | 2011-10-20 | | Release date: | 2012-03-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | N-Terminal Domain of Bombyx mori Fibroin Mediates the Assembly of Silk in Response to pH Decrease.
J.Mol.Biol., 418, 2012
|
|
5DRT
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD2 [2-(2-(5-((2-chlorophenoxy)methyl)-1H-tetrazol-1-yl)acetyl)-N-(4-chlorophenyl)hydrazinecarboxamide] | | Descriptor: | 2-({5-[(2-chlorophenoxy)methyl]-1H-tetrazol-1-yl}acetyl)-N-(4-chlorophenyl)hydrazinecarboxamide, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-16 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
5DSX
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD10 [6'-chloro-1,4-dimethyl-5'-(2-methyl-6-((4-(methylamino)pyrimidin-2-yl)amino)-1H-indol-1-yl)-[3,3'-bipyridin]-2(1H)-one] | | Descriptor: | 6'-chloro-1,4-dimethyl-5'-(2-methyl-6-{[4-(methylamino)pyrimidin-2-yl]amino}-1H-indol-1-yl)-3,3'-bipyridin-2(1H)-one, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-17 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
4OB8
 
 | | Crystal structure of a novel thermostable esterase from Pseudomonas putida ECU1011 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | Deposit date: | 2014-01-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
5HMM
 
 | | Crystal Structure of T5 D15 Protein Co-crystallized with Metal Ions | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exodeoxyribonuclease, ... | | Authors: | Flemming, C.S, Sedelnikova, S.E, Rafferty, J.B, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2016-01-16 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
7DLA
 
 | |
5XGH
 
 | | Crystal structure of PI3K complex with an inhibitor | | Descriptor: | 3-[(4-fluorophenyl)methylamino]-5-(4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenol, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Song, K, Yang, X, Zhao, Y, Jian, Z. | | Deposit date: | 2017-04-13 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | New Insights into PI3K Inhibitor Design using X-ray Structures of PI3K alpha Complexed with a Potent Lead Compound.
Sci Rep, 7, 2017
|
|
8IK2
 
 | | RhlA exhibits dual thioesterase and acyltransferase activities during rhamnolipid biosynthesis | | Descriptor: | (3~{S})-3-oxidanyldecanoic acid, 3-(3-hydroxydecanoyloxy)decanoate synthase | | Authors: | Tang, T, Fu, L.H, Xie, W.H, Luo, Y.Z, Zhang, Y.T, Si, T. | | Deposit date: | 2023-02-28 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | RhlA Exhibits Dual Thioesterase and Acyltransferase Activities during Rhamnolipid Biosynthesis
Acs Catalysis, 13, 2023
|
|
8JAL
 
 | | Structure of CRL2APPBP2 bound with RxxGP degron (dimer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, Elongin-B, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAR
 
 | | Structure of CRL2APPBP2 bound with RxxGPAA degron (dimer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, Elongin-B, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6IZG
 
 | |
8JAQ
 
 | | Structure of CRL2APPBP2 bound with RxxGP degron (tetramer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAS
 
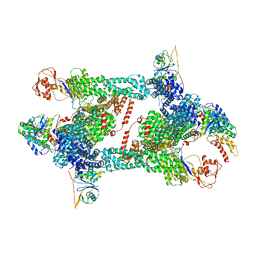 | | Structure of CRL2APPBP2 bound with RxxGPAA degron (tetramer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAU
 
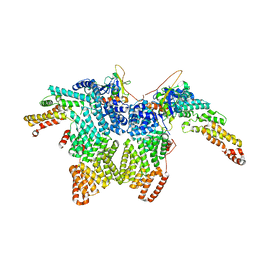 | | Structure of CRL2APPBP2 bound with the C-degron of MRPL28 (dimer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, Elongin-B, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAV
 
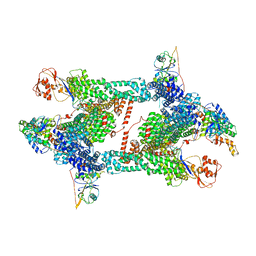 | | Structure of CRL2APPBP2 bound with the C-degron of MRPL28 (tetramer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4DHY
 
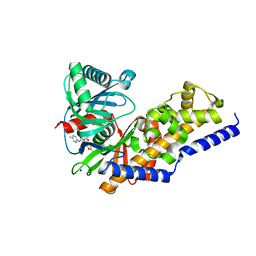 | | Crystal structure of human glucokinase in complex with glucose and activator | | Descriptor: | Glucokinase, N,N-dimethyl-5-({2-methyl-6-[(5-methylpyrazin-2-yl)carbamoyl]-1-benzofuran-4-yl}oxy)pyrimidine-2-carboxamide, SODIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Insights into Mechanism of Glucokinase Activation: OBSERVATION OF MULTIPLE DISTINCT PROTEIN CONFORMATIONS.
J.Biol.Chem., 287, 2012
|
|
7X08
 
 | | S protein of SARS-CoV-2 in complex with 2G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Broad ultra-potent neutralization of SARS-CoV-2 variants by monoclonal antibodies specific to the tip of RBD.
Cell Discov, 8, 2022
|
|
5YRN
 
 | | Structure of RIP2 CARD domain | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Wu, B, Gong, Q. | | Deposit date: | 2017-11-09 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of RIP2 activation and signaling.
Nat Commun, 9, 2018
|
|
7WWM
 
 | | S protein of Delta variant in complex with ZWC6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-13 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Broadly neutralizing antibodies against Omicron-included SARS-CoV-2 variants induced by vaccination.
Signal Transduct Target Ther, 7, 2022
|
|
7WWL
 
 | | S protein of Delta variant in complex with ZWD12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-13 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Broadly neutralizing antibodies against Omicron-included SARS-CoV-2 variants induced by vaccination.
Signal Transduct Target Ther, 7, 2022
|
|
5K08
 
 | | RecA mini intein-Zeise's salt complex | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, RecA mini intein, SULFATE ION, ... | | Authors: | Li, Z, Li, H.M. | | Deposit date: | 2016-05-17 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural insights into protein splicing inhibition by platinum therapeutics as potential anti-microbials
To be published
|
|
