6M6E
 
 | |
6M6F
 
 | |
8Z90
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription initiation conformation 2 | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z98
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription reception conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z8N
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 3 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z97
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription elongation conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z9H
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription elongation-reception conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z9Q
 
 | | Cryo-EM structure of Thogoto virus polymerase in a replication reception conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(ATP))-R(P*GP*CP*AP*AP*AP*AP*AP*CP*A)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z8J
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 2 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z9R
 
 | | Cryo-EM structure of Thogoto virus polymerase in a replication elongation-reception conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(ATP))-R(P*GP*CP*AP*AP*AP*AP*AP*CP*A)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z85
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 1 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-21 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z8X
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription initiation conformation | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8EX5
 
 | |
8EX6
 
 | | Human S1P transporter Spns2 in an inward-facing open conformation (state 1*) | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Ahmed, S, Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate.
Cell, 186, 2023
|
|
8EX4
 
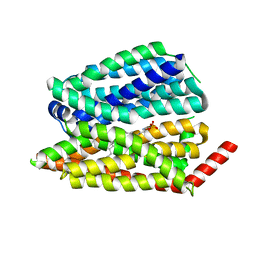 | | Human S1P transporter Spns2 in an inward-facing open conformation (state 1) | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Ahmed, S, Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate.
Cell, 186, 2023
|
|
8EX7
 
 | |
8EX8
 
 | |
8X3V
 
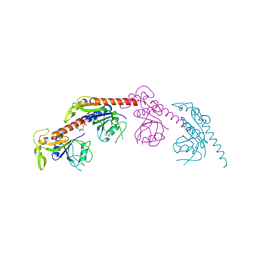 | |
4IOX
 
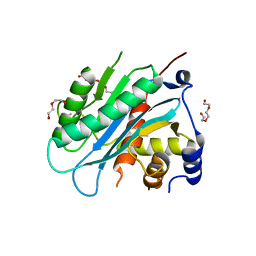 | | The structure of the herpes simplex virus DNA-packaging motor pUL15 C-terminal nuclease domain provides insights into cleavage of concatemeric viral genome precursors | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Selvarajan Sigamani, S, Zhao, H, Kamau, Y, Tang, L. | | Deposit date: | 2013-01-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.458 Å) | | Cite: | The Structure of the Herpes Simplex Virus DNA-Packaging Terminase pUL15 Nuclease Domain Suggests an Evolutionary Lineage among Eukaryotic and Prokaryotic Viruses.
J.Virol., 87, 2013
|
|
7YN9
 
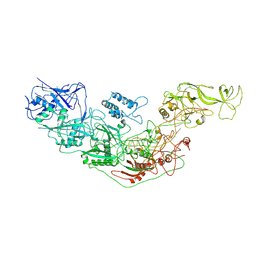 | | Cryo-EM structure of Cas7-11-crRNA binary complex | | Descriptor: | CRISPR-associated RAMP family protein, crRNA | | Authors: | Huo, Y, Dong, Q, Zhao, H, Jiang, T. | | Deposit date: | 2022-07-30 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structure and protease activity of the type III-E CRISPR-Cas effector.
Nat Microbiol, 8, 2023
|
|
7YNC
 
 | | Cryo-EM structure of Cas7-11-crRNA bound to target RNA-3 | | Descriptor: | CRISPR-associated RAMP family protein, Target RNA-3 (28-MER), crRNA (38-MER) | | Authors: | Huo, Y, Dong, Q, Zhao, H, Jiang, T. | | Deposit date: | 2022-07-30 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Cryo-EM structure and protease activity of the type III-E CRISPR-Cas effector.
Nat Microbiol, 8, 2023
|
|
7YNA
 
 | | Cryo-EM structure of Cas7-11-crRNA bound to target RNA-1 | | Descriptor: | CRISPR-associated RAMP family protein, Target RNA-1 (25-MER), crRNA (38-MER) | | Authors: | Huo, Y, Dong, Q, Zhao, H, Jiang, T. | | Deposit date: | 2022-07-30 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Cryo-EM structure and protease activity of the type III-E CRISPR-Cas effector.
Nat Microbiol, 8, 2023
|
|
7YNB
 
 | | Cryo-EM structure of Cas7-11-crRNA bound to target RNA-2 | | Descriptor: | CRISPR-associated RAMP family protein, Target RNA-2 (28-MER), crRNA (38-MER) | | Authors: | Huo, Y, Dong, Q, Zhao, H, Jiang, T. | | Deposit date: | 2022-07-30 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Cryo-EM structure and protease activity of the type III-E CRISPR-Cas effector.
Nat Microbiol, 8, 2023
|
|
7YND
 
 | | Cryo-EM structure of Cas7-11-crRNA-Csx29 ternary complex | | Descriptor: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, crRNA (38-MER) | | Authors: | Huo, Y, Dong, Q, Zhao, H, Jiang, T. | | Deposit date: | 2022-07-30 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structure and protease activity of the type III-E CRISPR-Cas effector.
Nat Microbiol, 8, 2023
|
|
4KW3
 
 | |
