1F56
 
 | | SPINACH PLANTACYANIN | | Descriptor: | COPPER (I) ION, PLANTACYANIN, SULFATE ION | | Authors: | Einsle, O, Mehrabian, Z, Nalbandyan, R, Messerschmidt, A. | | Deposit date: | 2000-06-13 | | Release date: | 2000-11-01 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of plantacyanin, a basic blue cupredoxin from spinach.
J.Biol.Inorg.Chem., 5, 2000
|
|
5AJ0
 
 | | Cryo electron microscopy of actively translating human polysomes (POST state). | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Behrmann, E, Loerke, J, Budkevich, T.V, Yamamoto, K, Schmidt, A, Penczek, P.A, Vos, M.R, Burger, J, Mielke, T, Scheerer, P, Spahn, C.M.T. | | Deposit date: | 2015-02-19 | | Release date: | 2015-05-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Snapshots of Actively Translating Human Ribosomes
Cell(Cambridge,Mass.), 161, 2015
|
|
5MDL
 
 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its O2-derivatized form by a "soak-and-freeze" derivatization method | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, ... | | Authors: | Kalms, J, Schmidt, A, Scheerer, P. | | Deposit date: | 2016-11-11 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Tracking the route of molecular oxygen in O2-tolerant membrane-bound [NiFe] hydrogenase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2UX7
 
 | | Pseudoazurin with engineered amicyanin ligand loop, reduced form, pH 7.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-27 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Influence of loop shortening on the metal binding site of cupredoxin pseudoazurin.
Biochemistry, 46, 2007
|
|
2UX6
 
 | | Pseudoazurin with engineered amicyanin ligand loop, oxidized form, pH 7.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-27 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Influence of Loop Shortening on the Metal Binding Site of Cupredoxin Pseudoazurin.
Biochemistry, 46, 2007
|
|
5D51
 
 | | Krypton derivatization of an O2-tolerant membrane-bound [NiFe] hydrogenase reveals a hydrophobic gas tunnel network | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Kalms, J, Schmidt, A, Frielingsdorf, S, van der Linden, P, von Stetten, D, Lenz, O, Carpentier, P, Scheerer, P. | | Deposit date: | 2015-08-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Krypton Derivatization of an O2 -Tolerant Membrane-Bound [NiFe] Hydrogenase Reveals a Hydrophobic Tunnel Network for Gas Transport.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2UXF
 
 | | Pseudoazurin with engineered amicyanin ligand loop, oxidized form, pH 5.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-28 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Influence of Loop Shortening on the Metal Binding Site of Cupredoxin Pseudoazurin.
Biochemistry, 46, 2007
|
|
2UXG
 
 | | Pseudoazurin with engineered amicyanin ligand loop, reduced form, pH 5.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-28 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Influence of Loop Shortening on the Metal Binding Site of Cupredoxin Pseudoazurin.
Biochemistry, 46, 2007
|
|
5FLX
 
 | | Mammalian 40S HCV-IRES complex | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Yamamoto, H, Collier, M, Loerke, J, Ismer, J, Schmidt, A, Hilal, T, Sprink, T, Yamamoto, K, Mielke, T, Burger, J, Shaikh, T.R, Dabrowski, M, Hildebrand, P.W, Scheerer, P, Spahn, C.M.T. | | Deposit date: | 2015-10-28 | | Release date: | 2015-12-23 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Architecture of the Ribosome-Bound Hepatitis C Virus Internal Ribosomal Entry Site RNA.
Embo J., 34, 2015
|
|
1POI
 
 | | CRYSTAL STRUCTURE OF GLUTACONATE COENZYME A-TRANSFERASE FROM ACIDAMINOCOCCUS FERMENTANS TO 2.55 ANGSTOMS RESOLUTION | | Descriptor: | COPPER (II) ION, GLUTACONATE COENZYME A-TRANSFERASE | | Authors: | Jacob, U, Mack, M, Clausen, T, Huber, R, Buckel, W, Messerschmidt, A. | | Deposit date: | 1997-01-24 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glutaconate CoA-transferase from Acidaminococcus fermentans: the crystal structure reveals homology with other CoA-transferases.
Structure, 5, 1997
|
|
1QDB
 
 | | CYTOCHROME C NITRITE REDUCTASE | | Descriptor: | CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, HEME C, ... | | Authors: | Einsle, O, Messerschmidt, A, Stach, P, Huber, R, Kroneck, P.M.H. | | Deposit date: | 1999-05-19 | | Release date: | 1999-08-18 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of cytochrome c nitrite reductase.
Nature, 400, 1999
|
|
1E5Z
 
 | |
1X9R
 
 | | Umecyanin from Horse Raddish- Crystal Structure of the oxidised form | | Descriptor: | COPPER (II) ION, Umecyanin | | Authors: | Koch, M, Velarde, M, Harrison, M.D, Echt, S, Fischer, M, Messerschmidt, A, Dennison, C. | | Deposit date: | 2004-08-24 | | Release date: | 2005-03-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Oxidized and Reduced Stellacyanin from Horseradish Roots
J.Am.Chem.Soc., 127, 2005
|
|
1QGN
 
 | | CYSTATHIONINE GAMMA-SYNTHASE FROM NICOTIANA TABACUM | | Descriptor: | PROTEIN (CYSTATHIONINE GAMMA-SYNTHASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Steegborn, C, Messerschmidt, A, Laber, B, Streber, W, Huber, R, Clausen, T. | | Deposit date: | 1999-05-02 | | Release date: | 1999-08-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of cystathionine gamma-synthase from Nicotiana tabacum reveals its substrate and reaction specificity.
J.Mol.Biol., 290, 1999
|
|
1E5Y
 
 | |
1XDW
 
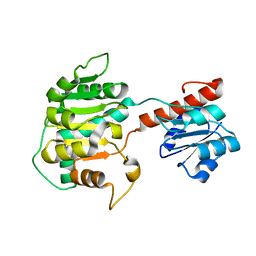 | | NAD+-dependent (R)-2-Hydroxyglutarate Dehydrogenase from Acidaminococcus fermentans | | Descriptor: | NAD+-dependent (R)-2-Hydroxyglutarate Dehydrogenase | | Authors: | Martins, B.M, Macedo-Ribeiro, S, Bresser, J, Buckel, W, Messerschmidt, A. | | Deposit date: | 2004-09-08 | | Release date: | 2005-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for stereo-specific catalysis in NAD(+)-dependent (R)-2-hydroxyglutarate dehydrogenase from Acidaminococcus fermentans.
Febs J., 272, 2005
|
|
8AZW
 
 | | Cryo-EM structure of the plant 60S subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | Deposit date: | 2022-09-06 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8AUV
 
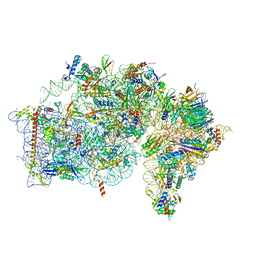 | | Cryo-EM structure of the plant 40S subunit | | Descriptor: | 18S rRNA, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
1SEZ
 
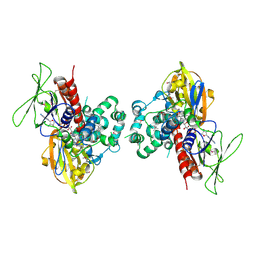 | | Crystal Structure of Protoporphyrinogen IX Oxidase | | Descriptor: | 2-{2-[4-(1,1,3,3-TETRAMETHYLBUTYL)PHENOXY]ETHOXY}ETHANOL, 4-BROMO-3-(5'-CARBOXY-4'-CHLORO-2'-FLUOROPHENYL)-1-METHYL-5-TRIFLUOROMETHYL-PYRAZOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Koch, M, Breithaupt, C, Kiefersauer, R, Freigang, J, Huber, R, Messerschmidt, A. | | Deposit date: | 2004-02-19 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of protoporphyrinogen IX oxidase: a key enzyme in haem and chlorophyll biosynthesis.
Embo J., 23, 2004
|
|
8B2L
 
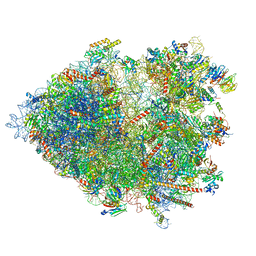 | | Cryo-EM structure of the plant 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 30S ribosomal protein S15, ... | | Authors: | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | Deposit date: | 2022-09-14 | | Release date: | 2023-08-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
1X9U
 
 | | Umecyanin from Horse Raddish- Crystal Structure of the reduced form | | Descriptor: | COPPER (II) ION, Umecyanin | | Authors: | Koch, M, Velarde, M, Harrison, M.D, Echt, S, Fischer, M, Messerschmidt, A, Dennison, C. | | Deposit date: | 2004-08-24 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Oxidized and Reduced Stellacyanin from Horseradish Roots
J.Am.Chem.Soc., 127, 2005
|
|
2AZU
 
 | |
4IBL
 
 | | Rubidium Sites in Blood Coagulation Factor VIIa | | Descriptor: | BENZAMIDINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vadivel, K, Schmidt, A, Cascio, D, Padmanabhan, K, Bajaj, S.P. | | Deposit date: | 2012-12-08 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human factor VIIa-soluble tissue factor with calcium, magnesium and rubidium
Acta Crystallogr.,Sect.D, D77, 2021
|
|
4J26
 
 | | Estrogen Receptor in complex with proline-flanked LXXLL peptides | | Descriptor: | 12-mer Peptide, ESTRADIOL, Estrogen receptor beta | | Authors: | Fuchs, S, Nguyen, H.D, Phan, T, Burton, M, Nieto, L, de Vries-van Leeuwen, I, Schmidt, A, Goodarzifard, M, Agten, S, Rose, R, Ottmann, C, Milroy, L.G, Brunsveld, L. | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Proline primed helix length as a modulator of the nuclear receptor-coactivator interaction
J.Am.Chem.Soc., 135, 2013
|
|
4J24
 
 | | Estrogen Receptor in complex with proline-flanked LXXLL peptides | | Descriptor: | 19-mer peptide, ESTRADIOL, Estrogen receptor beta | | Authors: | Fuchs, S, Nguyen, H.D, Phan, T, Burton, M, Nieto, L, de Vries-van Leeuwen, I, Schmidt, A, Goodarzifard, M, Agten, S, Rose, R, Ottmann, C, Milroy, L.G, Brunsveld, L. | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Proline primed helix length as a modulator of the nuclear receptor-coactivator interaction
J.Am.Chem.Soc., 135, 2013
|
|
