7EVT
 
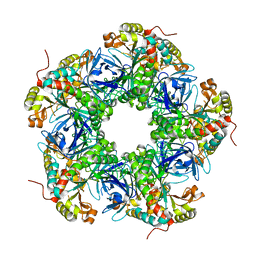 | | Crystal structure of the N-terminal degron-truncated human glutamine synthetase | | Descriptor: | Glutamine synthetase | | Authors: | Chek, M.F, Kim, S.Y, Mori, T, Hakoshima, T. | | Deposit date: | 2021-05-22 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of N-terminal degron-truncated human glutamine synthetase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
2E1E
 
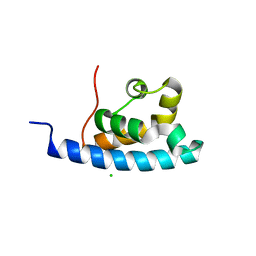 | | Crystal structure of the HRDC Domain of Human Werner Syndrome Protein, WRN | | Descriptor: | CHLORIDE ION, Werner syndrome ATP-dependent helicase | | Authors: | Kitano, K, Yoshihara, N, Hakoshima, T. | | Deposit date: | 2006-10-25 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the HRDC domain of human Werner syndrome protein, WRN
J.Biol.Chem., 282, 2007
|
|
2F2U
 
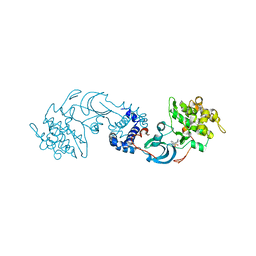 | | crystal structure of the Rho-kinase kinase domain | | Descriptor: | 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, Rho-associated protein kinase 2 | | Authors: | Yamaguchi, H, Hakoshima, T. | | Deposit date: | 2005-11-18 | | Release date: | 2006-04-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism for the regulation of rho-kinase by dimerization and its inhibition by fasudil
Structure, 14, 2006
|
|
2H9V
 
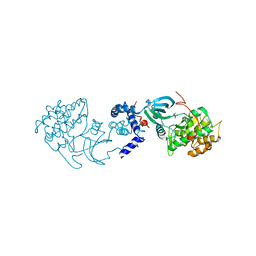 | | Structural basis for induced-fit binding of Rho-kinase to the inhibitor Y27632 | | Descriptor: | (R)-TRANS-4-(1-AMINOETHYL)-N-(4-PYRIDYL) CYCLOHEXANECARBOXAMIDE, Rho-associated protein kinase 2 | | Authors: | Yamaguchi, H, Miwa, Y, Kasa, M, Kitano, K, Amano, M, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2006-06-12 | | Release date: | 2006-12-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for induced-fit binding of Rho-kinase to the inhibitor Y-27632
J.Biochem.(Tokyo), 140, 2006
|
|
1IS8
 
 | | Crystal structure of rat GTPCHI/GFRP stimulatory complex plus Zn | | Descriptor: | GTP Cyclohydrolase I, GTP Cyclohydrolase I Feedback Regulatory Protein, PHENYLALANINE, ... | | Authors: | Maita, N, Okada, K, Hatakeyama, K, Hakoshima, T. | | Deposit date: | 2001-11-18 | | Release date: | 2002-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the stimulatory complex of GTP cyclohydrolase I and its feedback regulatory protein GFRP.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IS7
 
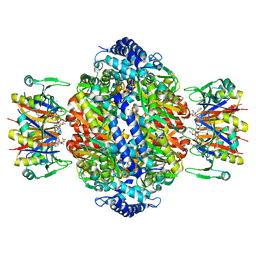 | | Crystal structure of rat GTPCHI/GFRP stimulatory complex | | Descriptor: | GTP Cyclohydrolase I, GTP Cyclohydrolase I Feedback Regulatory Protein, PHENYLALANINE, ... | | Authors: | Maita, N, Okada, K, Hatakeyama, K, Hakoshima, T. | | Deposit date: | 2001-11-18 | | Release date: | 2002-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the stimulatory complex of GTP cyclohydrolase I and its feedback regulatory protein GFRP.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2ZSI
 
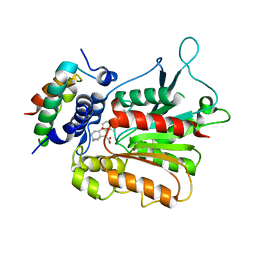 | | Structural basis of gibberellin(GA4)-induced DELLA recognition by the gibberellin receptor | | Descriptor: | DELLA protein GAI, GIBBERELLIN A4, Probable gibberellin receptor GID1L1 | | Authors: | Murase, K, Hirano, Y, Sun, T.P, Hakoshima, T. | | Deposit date: | 2008-09-10 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gibberellin-induced DELLA recognition by the gibberellin receptor GID1
Nature, 456, 2008
|
|
2ZSH
 
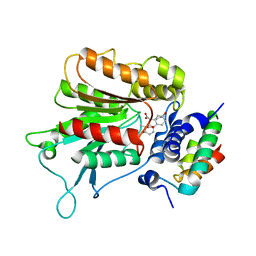 | | Structural basis of gibberellin(GA3)-induced DELLA recognition by the gibberellin receptor | | Descriptor: | DELLA protein GAI, GIBBERELLIN A3, Probable gibberellin receptor GID1L1 | | Authors: | Murase, K, Hirano, Y, Sun, T.P, Hakoshima, T. | | Deposit date: | 2008-09-10 | | Release date: | 2008-11-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gibberellin-induced DELLA recognition by the gibberellin receptor GID1
Nature, 456, 2008
|
|
3AAF
 
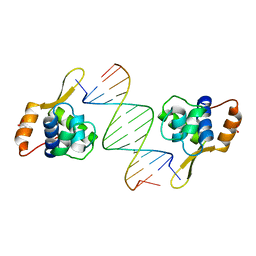 | | Structure of WRN RQC domain bound to double-stranded DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*AP*CP*CP*CP*TP*AP*AP*TP*TP*AP*GP*GP*GP*T)-3'), Werner syndrome ATP-dependent helicase | | Authors: | Kitano, K, Hakoshima, T. | | Deposit date: | 2009-11-16 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN
Structure, 18, 2010
|
|
336D
 
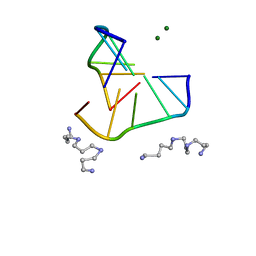 | | INTERACTION BETWEEN LEFT-HANDED Z-DNA AND POLYAMINE-3 THE CRYSTAL STRUCTURE OF THE D(CG)3 AND THERMOSPERMINE COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, N-(3-AMINO-PROPYL)-N-(5-AMINOPROPYL)-1,4-DIAMINOBUTANE | | Authors: | Ohishi, H, Terasoma, N, Nakanishi, I, Van Der Marel, G, Van Boom, J.H, Rich, A, Wang, A.H.-J, Hakoshima, T, Tomita, K.-I. | | Deposit date: | 1997-06-24 | | Release date: | 1998-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Interaction between left-handed Z-DNA and polyamine - 3. The crystal structure of the d(CG)3 and thermospermine complex.
FEBS Lett., 398, 1996
|
|
3W9R
 
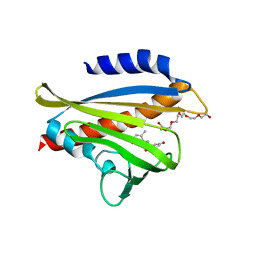 | | Crystal structure of the high-affinity abscisic acid receptor PYL9/RCAR9 bound to ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL9, HEXAETHYLENE GLYCOL | | Authors: | Nakagawa, M, Hirano, Y, Kagiyama, M, Shibata, N, Hakoshima, T. | | Deposit date: | 2013-04-13 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of high-affinity abscisic acid binding to PYL9/RCAR1.
Genes Cells, 19, 2014
|
|
3WE2
 
 | |
1WPL
 
 | | Crystal structure of the inhibitory form of rat GTP cyclohydrolase I/GFRP complex | | Descriptor: | 7,8-DIHYDROBIOPTERIN, GTP cyclohydrolase I, GTP cyclohydrolase I feedback regulatory protein, ... | | Authors: | Maita, N, Hatakeyama, K, Okada, K, Hakoshima, T. | | Deposit date: | 2004-09-08 | | Release date: | 2004-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of biopterin-induced inhibition of GTP cyclohydrolase I by GFRP, its feedback regulatory protein
J.Biol.Chem., 279, 2004
|
|
1WE0
 
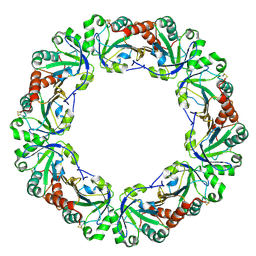 | | Crystal structure of peroxiredoxin (AhpC) from Amphibacillus xylanus | | Descriptor: | AMMONIUM ION, alkyl hydroperoxide reductase C | | Authors: | Kitano, K, Kita, A, Hakoshima, T, Niimura, Y, Miki, K. | | Deposit date: | 2004-05-21 | | Release date: | 2005-03-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of decameric peroxiredoxin (AhpC) from Amphibacillus xylanus
Proteins, 59, 2005
|
|
1VTW
 
 | | AT Base Pairs Are Less Stable than GC Base Pairs in Z-DNA: The Crystal Structure of D(M(5)CGTAM(5)CG) | | Descriptor: | DNA (5'-D(*(CH3)CP*GP*TP*AP*(CH3)CP*G)-3') | | Authors: | Wang, A.H.-J, Hakoshima, T, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | AT Base Pairs Are Less Stable than GC Base Pairs in Z-DNA: The Crystal Structure of d(m(5)CGTAm(5)CG)
Cell(Cambridge,Mass.), 37, 1984
|
|
1VTG
 
 | | THE MOLECULAR STRUCTURE OF A DNA-TRIOSTIN A COMPLEX | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), triostin A | | Authors: | Wang, A.H.-J, Ughetto, G, Quigley, G.J, Hakoshima, T, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The molecular structure of a DNA-triostin A complex.
Science, 225, 1984
|
|
1UL1
 
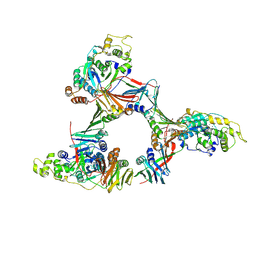 | | Crystal structure of the human FEN1-PCNA complex | | Descriptor: | Flap endonuclease-1, MAGNESIUM ION, Proliferating cell nuclear antigen | | Authors: | Sakurai, S, Kitano, K, Yamaguchi, H, Hamada, K, Okada, K, Fukuda, K, Uchida, M, Ohtsuka, E, Morioka, H, Hakoshima, T. | | Deposit date: | 2003-09-05 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for recruitment of human flap endonuclease 1 to PCNA
EMBO J., 24, 2005
|
|
6A1C
 
 | | Crystal structure of the CK2a1-go289 complex | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-4-[(E)-(3-methylsulfanyl-5-phenyl-1,2,4-triazol-4-yl)iminomethyl]phenol, Casein kinase II subunit alpha, ... | | Authors: | Kinoshita, T, Tsuyuguchi, M. | | Deposit date: | 2018-06-07 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Cell-based screen identifies a new potent and highly selective CK2 inhibitor for modulation of circadian rhythms and cancer cell growth.
Sci Adv, 5, 2019
|
|
1IDM
 
 | | 3-ISOPROPYLMALATE DEHYDROGENASE, LOOP-DELETED CHIMERA | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Sakurai, M, Ohzeki, M, Moriyama, H, Sato, M, Tanaka, N. | | Deposit date: | 1995-05-19 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a loop-deleted variant of 3-isopropylmalate dehydrogenase from Thermus thermophilus: an internal reprieve tolerance mechanism.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1XAB
 
 | |
1XAA
 
 | |
1XAC
 
 | |
1XAD
 
 | |
6LUE
 
 | | Crystal structure of mouse Cryptochrome 1 in complex with compound KL201 | | Descriptor: | 2-bromanyl-N-(5,6,7,8-tetrahydro-[1]benzothiolo[2,3-d]pyrimidin-4-yl)benzamide, Cryptochrome-1 | | Authors: | Miller, S, Aikawa, Y, Hirota, T. | | Deposit date: | 2020-01-27 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Isoform-Selective Modulator of Cryptochrome 1 Regulates Circadian Rhythms in Mammals.
Cell Chem Biol, 27, 2020
|
|
5XNF
 
 | | Crystal structure of the branched-chain polyamine synthase (BpsA) from Thermococcus kodakarensis | | Descriptor: | FE (III) ION, GLYCEROL, N(4)-bis(aminopropyl)spermidine synthase, ... | | Authors: | Mizohata, E, Tse, K.M, Fujita, J, Inoue, T. | | Deposit date: | 2017-05-22 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active site geometry of a novel aminopropyltransferase for biosynthesis of hyperthermophile-specific branched-chain polyamine.
FEBS J., 284, 2017
|
|
