1UC5
 
 | | Structure of diol dehydratase complexed with (R)-1,2-propanediol | | Descriptor: | AMMONIUM ION, CYANOCOBALAMIN, POTASSIUM ION, ... | | Authors: | Shibata, N, Nakanishi, Y, Fukuoka, M, Yamanishi, M, Yasuoka, N, Toraya, T. | | Deposit date: | 2003-04-08 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural rationalization for the lack of stereospecificity in coenzyme B12-dependent diol dehydratase
J.Biol.Chem., 278, 2003
|
|
5X9A
 
 | | Crystal structure of calaxin with calcium | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calaxin | | Authors: | Shojima, T, Hou, F, Takahashi, Y, Okai, M, Mizuno, K, Inaba, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2017-03-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a Ca2+-dependent regulator of flagellar motility reveals the open-closed structural transition
Sci Rep, 8, 2018
|
|
2RSY
 
 | | Solution structure of the SH2 domain of Csk in complex with a phosphopeptide from Cbp | | Descriptor: | Phosphoprotein associated with glycosphingolipid-enriched microdomains 1, Tyrosine-protein kinase CSK | | Authors: | Tanaka, H, Akagi, K, Oneyama, C, Tanaka, M, Sasaki, Y, Kanou, T, Lee, Y, Yokogawa, D, Debenecker, M, Nakagawa, A, Okada, M, Ikegami, T. | | Deposit date: | 2012-09-10 | | Release date: | 2013-04-10 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Identification of a new interaction mode between the Src homology 2 domain of C-terminal Src kinase (Csk) and Csk-binding protein/phosphoprotein associated with glycosphingolipid microdomains.
J.Biol.Chem., 288, 2013
|
|
3A3V
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase Y198F mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | Authors: | Hidaka, M, Fushinobu, S, Honda, Y, Kitaoka, M. | | Deposit date: | 2009-06-22 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 147, 2010
|
|
2DEP
 
 | |
3QFZ
 
 | | Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cellobiose Phosphorylase, ... | | Authors: | Fushinobu, S, Hidaka, M, Hayashi, A.M, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2011-01-24 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Interactions between glycoside hydrolase family 94 cellobiose phosphorylase and glucosidase inhibitors
J.Appl.Glyosci., 58, 2011
|
|
3DMI
 
 | | Crystallization and Structural Analysis of Cytochrome c6 from the Diatom Phaeodactylum tricornutum at 1.5 A resolution | | Descriptor: | HEME C, MAGNESIUM ION, cytochrome c6 | | Authors: | Akazaki, H, Kawai, F, Hosokawa, M, Hama, T, Hirano, T, Lim, B.-K, Sakurai, N, Hakamata, W, Park, S.-Y, Nishio, T, Oku, T. | | Deposit date: | 2008-07-01 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallization and structural analysis of cytochrome c(6) from the diatom Phaeodactylum tricornutum at 1.5 A resolution.
Biosci.Biotechnol.Biochem., 73, 2009
|
|
3QFY
 
 | | Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, Cellobiose Phosphorylase, ... | | Authors: | Fushinobu, S, Hidaka, M, Hayashi, A.M, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2011-01-24 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Interactions between glycoside hydrolase family 94 cellobiose phosphorylase and glucosidase inhibitors
J.Appl.Glyosci., 58, 2011
|
|
3QG0
 
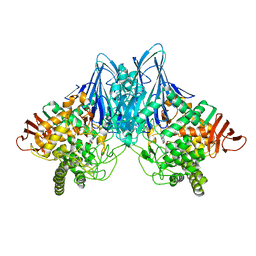 | | Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, Cellobiose Phosphorylase, PHOSPHATE ION, ... | | Authors: | Fushinobu, S, Hidaka, M, Hayashi, A.M, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2011-01-24 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interactions between glycoside hydrolase family 94 cellobiose phosphorylase and glucosidase inhibitors
J.Appl.Glyosci., 58, 2011
|
|
5X6U
 
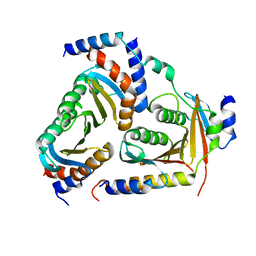 | | Crystal structure of human heteropentameric complex | | Descriptor: | Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, Ragulator complex protein LAMTOR3, ... | | Authors: | Yonehara, R, Nada, S, Nakai, T, Nakai, M, Kitamura, A, Ogawa, A, Nakatsumi, H, Nakayama, K.I, Li, S, Standley, D.M, Yamashita, E, Nakagawa, A, Okada, M. | | Deposit date: | 2017-02-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the assembly of the Ragulator-Rag GTPase complex.
Nat Commun, 8, 2017
|
|
1V7V
 
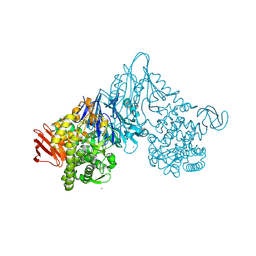 | | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase | | Descriptor: | CALCIUM ION, chitobiose phosphorylase | | Authors: | Hidaka, M, Honda, Y, Nirasawa, S, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Structure, 12, 2004
|
|
1VBH
 
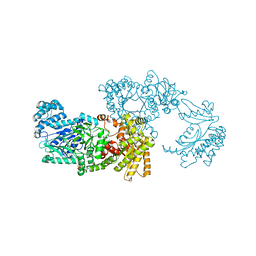 | | Pyruvate Phosphate Dikinase with bound Mg-PEP from Maize | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE, SULFATE ION, ... | | Authors: | Nakanishi, T, Nakatsu, T, Matsuoka, M, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of pyruvate phosphate dikinase from maize revealed an alternative conformation in the swiveling-domain motion
Biochemistry, 44, 2005
|
|
1V7W
 
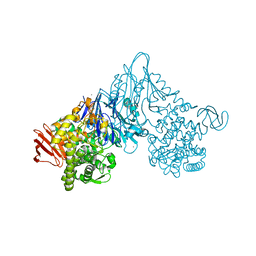 | | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Hidaka, M, Honda, Y, Nirasawa, S, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Structure, 12, 2004
|
|
1V7X
 
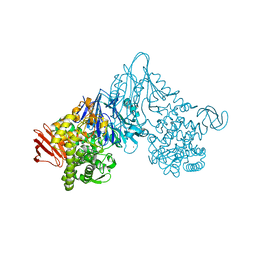 | | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc and sulfate | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Hidaka, M, Honda, Y, Nirasawa, S, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Structure, 12, 2004
|
|
1VBG
 
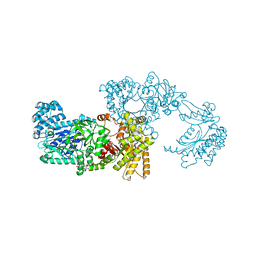 | | Pyruvate Phosphate Dikinase from Maize | | Descriptor: | MAGNESIUM ION, SULFATE ION, pyruvate,orthophosphate dikinase | | Authors: | Nakanishi, T, Nakatsu, T, Matsuoka, M, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of pyruvate phosphate dikinase from maize revealed an alternative conformation in the swiveling-domain motion
Biochemistry, 44, 2005
|
|
5B36
 
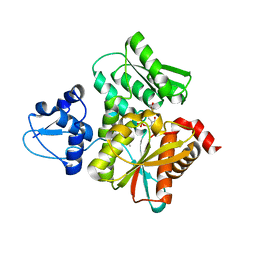 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with Cysteine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYSTEINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nakamura, T, Takeda, E, Kawai, Y, Kataoka, M, Ishikawa, K. | | Deposit date: | 2016-02-10 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Role of F225 in O-phosphoserine sulfhydrylase from Aeropyrum pernix K1
Extremophiles, 20, 2016
|
|
5H4J
 
 | | Crystal structure of Human dUTPase in complex with N-[(1R)-1-[3-(Cyclopentyloxy)-phenyl]-ethyl]-3-[(3,4-dihydro-2,4-dioxo-1(2H)-pyrimidinyl)methoxy]-1-propanesulfonamide | | Descriptor: | DIMETHYL SULFOXIDE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial, ... | | Authors: | Chong, K.T, Miyahara, S, Miyakoshi, H, Fukuoka, M. | | Deposit date: | 2016-11-01 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | TAS-114, a First-in-Class Dual dUTPase/DPD Inhibitor, Demonstrates Potential to Improve Therapeutic Efficacy of Fluoropyrimidine-Based Chemotherapy.
Mol. Cancer Ther., 17, 2018
|
|
4YPJ
 
 | | X-ray Structure of The Mutant of Glycoside Hydrolase | | Descriptor: | Beta galactosidase | | Authors: | Ishikawa, K, Kataoka, M, Yanamoto, T, Nakabayashi, M, Watanabe, M. | | Deposit date: | 2015-03-13 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of beta-galactosidase from Bacillus circulans ATCC 31382 (BgaD) and the construction of the thermophilic mutants.
Febs J., 282, 2015
|
|
6KU3
 
 | | Crystal structure of gibberellin 2-oxidase3 (GA2ox3)in rice | | Descriptor: | 2-OXOGLUTARIC ACID, GIBBERELLIN A4, GLYCEROL, ... | | Authors: | Takehara, S, Mikami, B, Sakuraba, S, Matsuoka, M, Ueguchi-Tanaka, M. | | Deposit date: | 2019-08-30 | | Release date: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A common allosteric mechanism regulates homeostatic inactivation of auxin and gibberellin.
Nat Commun, 11, 2020
|
|
6KUN
 
 | | Crystal structure of dioxygenase for auxin oxidation (DAO) in rice | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent dioxygenase DAO, ... | | Authors: | Takehara, S, Mikami, B, Sakuraba, S, Matsuoka, M, Ueguchi-Tanaka, M. | | Deposit date: | 2019-09-02 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | A common allosteric mechanism regulates homeostatic inactivation of auxin and gibberellin.
Nat Commun, 11, 2020
|
|
4Q0S
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase in complex with ribitol | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), D-ribitol, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4Q0P
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase in complex with L-ribose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4Q0V
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase mutant E204Q in complex with L-ribulose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4Q0Q
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase in complex with L-ribulose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4Q0U
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase mutant E204Q in complex with L-ribose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
