8A0H
 
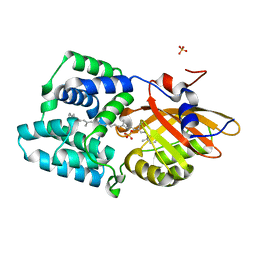 | | Crystal structure of the E25A mutant of the Orange Carotenoid Protein X from Gloeobacter kilaueensis JS1 complexed with echinenone | | Descriptor: | OCP N-terminal domain-containing protein, SULFATE ION, beta,beta-caroten-4-one | | Authors: | Boyko, K.M, Slonimskiy, Y.B, Zupnik, A.O, Varfolomeeva, L.A, Maksimov, E.G, Sluchanko, N.N. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A primordial Orange Carotenoid Protein: Structure, photoswitching activity and evolutionary aspects.
Int.J.Biol.Macromol., 222, 2022
|
|
8AH2
 
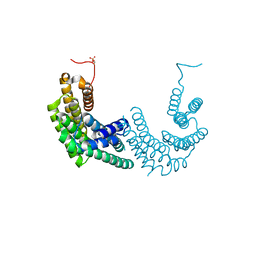 | | Crystal structure of human 14-3-3 zeta fused to the NPM1 peptide including phosphoserine-48 | | Descriptor: | 14-3-3 protein zeta/delta,Nucleophosmin | | Authors: | Boyko, K.M, Kapitonova, A.A, Tugaeva, K.V, Varfolomeeva, L.A, Sluchanko, N.N. | | Deposit date: | 2022-07-20 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the recognition by 14-3-3 proteins of a conditional binding site within the oligomerization domain of human nucleophosmin.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
8AAQ
 
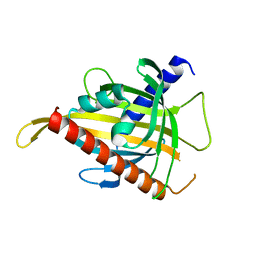 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP), CRT-416 form | | Descriptor: | Carotenoid-binding protein | | Authors: | Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Minyaev, M.E, Faletrov, Y.V, Boyko, K.M, Sluchanko, N.N. | | Deposit date: | 2022-07-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Preparation and Structural Studies of the Silkworm Carotenoid-Binding Protein Complexed with a New Pigment
Crystallography Reports, 2022
|
|
8C18
 
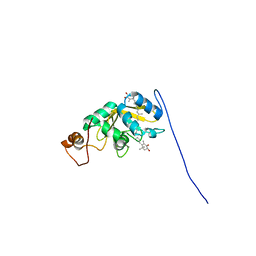 | | Solution structure of carotenoid-binding protein AstaPo1 in complex with astaxanthin | | Descriptor: | ASTAXANTHIN, Astaxanthin binding fasciclin family protein | | Authors: | Kornilov, F.D, Savitskaya, A.G, Slonimskiy, Y.B, Goncharuk, S.A, Sluchanko, N.N, Mineev, K.S. | | Deposit date: | 2022-12-20 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the ligand promiscuity of the neofunctionalized, carotenoid-binding fasciclin domain protein AstaP.
Commun Biol, 6, 2023
|
|
6T6O
 
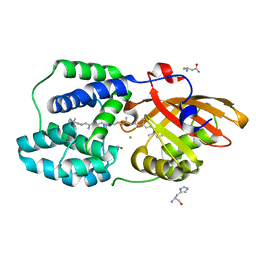 | | Y201W mutant of the orange carotenoid protein from Synechocystis at pH 4.6 | | Descriptor: | ASPARAGINE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sluchanko, N.N, Gushchin, I, Botnarevskiy, V.S, Slonimskiy, Y.B, Remeeva, A, Kovalev, K, Stepanov, A.V, Gordeliy, V, Maksimov, E.G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Role of hydrogen bond alternation and charge transfer states in photoactivation of the Orange Carotenoid Protein.
Commun Biol, 4, 2021
|
|
6T80
 
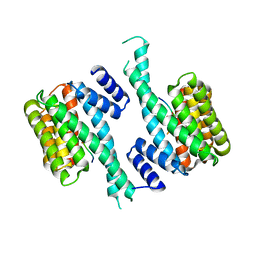 | |
7OAR
 
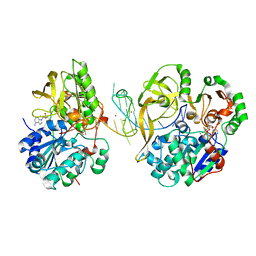 | | Crystal structure of helicase Pif1 from Thermus oshimai in complex with parallel G-quadruplex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (28-MER), MAGNESIUM ION, ... | | Authors: | Dai, Y.X, Liu, N.N, Guo, H.L, Chen, W.F, Rety, S, Xi, X.G. | | Deposit date: | 2021-04-20 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural mechanism underpinning Thermus oshimai Pif1-mediated G-quadruplex unfolding.
Embo Rep., 23, 2022
|
|
7Q16
 
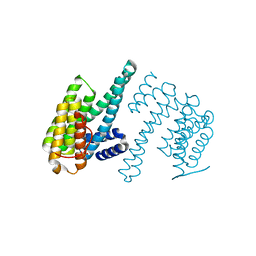 | | Human 14-3-3 zeta fused to the BAD peptide including phosphoserine-74 | | Descriptor: | 14-3-3 protein zeta/delta,Bcl2-associated agonist of cell death | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Gushchin, I, Remeeva, A, Kovalev, K, Cooley, R.B. | | Deposit date: | 2021-10-18 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Crystal structure of human 14-3-3 zeta complexed with the noncanonical phosphopeptide from proapoptotic BAD.
Biochem.Biophys.Res.Commun., 583, 2021
|
|
6T5H
 
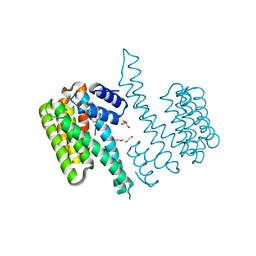 | | Human 14-3-3 sigma fused to the StARD1 peptide including phosphoserine-57 | | Descriptor: | 14-3-3 protein sigma,Steroidogenic acute regulatory protein, mitochondrial, IMIDAZOLE, ... | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Titterington, J, Antson, A.A. | | Deposit date: | 2019-10-16 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Molecular basis for the recognition of steroidogenic acute regulatory protein by the 14-3-3 protein family.
Febs J., 287, 2020
|
|
6TWZ
 
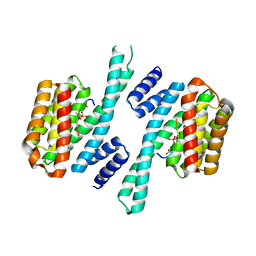 | | 14-3-3 sigma complexed with a phosphorylated 16E6 peptide | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D(-)-TARTARIC ACID, ... | | Authors: | Gogl, G, Cousido-Siah, A, Sluchanko, N.N, Trave, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual Specificity PDZ- and 14-3-3-Binding Motifs: A Structural and Interactomics Study.
Structure, 28, 2020
|
|
6T5F
 
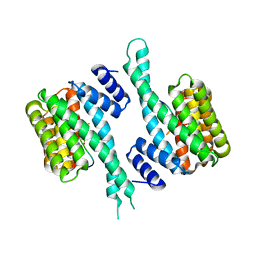 | | Human 14-3-3 sigma fused to the StARD1 peptide including phosphoserine-195 | | Descriptor: | 14-3-3 protein sigma, StARD1 peptide | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Titterington, J, Antson, A.A. | | Deposit date: | 2019-10-16 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Molecular basis for the recognition of steroidogenic acute regulatory protein by the 14-3-3 protein family.
Febs J., 287, 2020
|
|
5YEC
 
 | | Crystal structure of Atg7CTD-Atg8-MgATP complex in form II | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Autophagy-related protein 8, MAGNESIUM ION, ... | | Authors: | Yamaguchi, M, Satoo, K, Noda, N.N. | | Deposit date: | 2017-09-16 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Atg7 Activates an Autophagy-Essential Ubiquitin-like Protein Atg8 through Multi-Step Recognition.
J. Mol. Biol., 430, 2018
|
|
5W2F
 
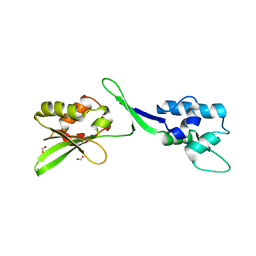 | | Crystal Structure of the C-terminal Domain of Human eIF2D at 1.4 A resolution | | Descriptor: | Eukaryotic translation initiation factor 2D, FORMIC ACID | | Authors: | Vaidya, A.T, Lomakin, I.B, Joseph, N.N, Dmitriev, S.E, Steitz, T.A. | | Deposit date: | 2017-06-06 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the C-terminal Domain of Human eIF2D and Its Implications on Eukaryotic Translation Initiation.
J. Mol. Biol., 429, 2017
|
|
2ZPN
 
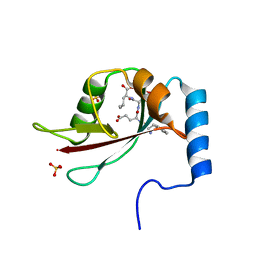 | |
3A7P
 
 | |
7W36
 
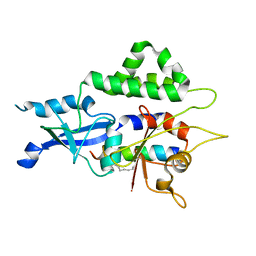 | |
3A77
 
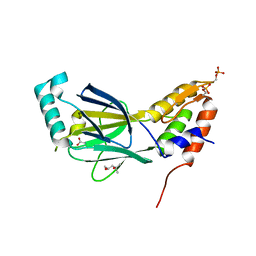 | | The crystal structure of phosphorylated IRF-3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Interferon regulatory factor 3 | | Authors: | Takahasi, K, Horiuchi, M, Noda, N.N, Inagaki, F. | | Deposit date: | 2009-09-17 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ser386 phosphorylation of transcription factor IRF-3 induces dimerization and association with CBP/p300 without overall conformational change.
Genes Cells, 15, 2010
|
|
3A7O
 
 | |
7YDO
 
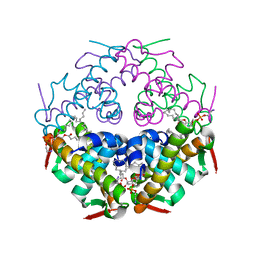 | | Crystal structure of Atg44 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Uncharacterized protein C26A3.14c | | Authors: | Maruyama, T, Noda, N.N. | | Deposit date: | 2022-07-04 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The mitochondrial intermembrane space protein mitofissin drives mitochondrial fission required for mitophagy.
Mol.Cell, 83, 2023
|
|
7W3O
 
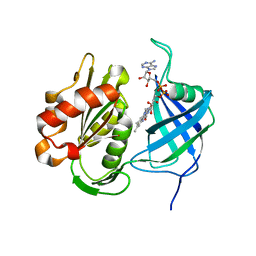 | | Crystal structure of human CYB5R3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3 soluble form | | Authors: | Noda, N.N. | | Deposit date: | 2021-11-25 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The UFM1 system regulates ER-phagy through the ufmylation of CYB5R3.
Nat Commun, 13, 2022
|
|
7W3N
 
 | | Crystal structure of Ufm1 fused to UFBP1 UFIM | | Descriptor: | UFBP1 peptide,Ubiquitin-fold modifier 1 | | Authors: | Noda, N.N. | | Deposit date: | 2021-11-25 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The UFM1 system regulates ER-phagy through the ufmylation of CYB5R3.
Nat Commun, 13, 2022
|
|
3CBR
 
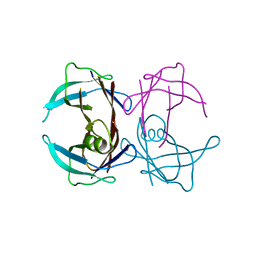 | | Crystal structure of human Transthyretin (TTR) at pH3.5 | | Descriptor: | Transthyretin | | Authors: | Mohamedmohaideen, N.N, Palaninathan, S.K, Snee, W.C, Kelly, J.W, C Sacchettini, J. | | Deposit date: | 2008-02-22 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into pH-induced conformational changes within the native human transthyretin tetramer.
J.Mol.Biol., 382, 2008
|
|
7W68
 
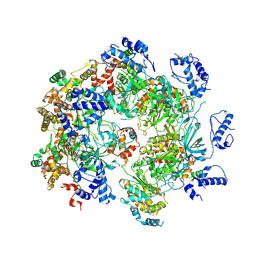 | | human single hexameric Mcm2-7 complex | | Descriptor: | DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, DNA replication licensing factor MCM4, ... | | Authors: | Xu, N.N, Lin, Q.P, Liu, C.D, Tian, H.L, Xiang, Y, Zhu, G. | | Deposit date: | 2021-12-01 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | human single hexameric Mcm2-7 complex
To Be Published
|
|
2ZZP
 
 | |
7YO8
 
 | |
