5JGP
 
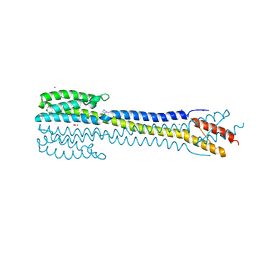 | | Crystal structure of the nitrate/nitrite sensor NarQ fragment bound with iodide ions | | Descriptor: | IODIDE ION, NITRATE ION, Nitrate/nitrite sensor protein NarQ | | Authors: | Melnikov, I, Polovinkin, V, Popov, A, Gordeliy, V. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fast iodide-SAD phasing for high-throughput membrane protein structure determination.
Sci Adv, 3, 2017
|
|
5JTB
 
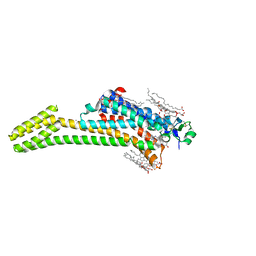 | | Crystal structure of the chimeric protein of A2aAR-BRIL with bound iodide ions | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Melnikov, I, Polovinkin, V, Shevtsov, M, Borshchevskiy, V, Cherezov, V, Popov, A, Gordeliy, V. | | Deposit date: | 2016-05-09 | | Release date: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fast iodide-SAD phasing for high-throughput membrane protein structure determination.
Sci Adv, 3, 2017
|
|
4XTL
 
 | | Crystal structure of the light-driven sodium pump KR2 in the monomeric blue form, pH 4.3 | | Descriptor: | EICOSANE, GLYCEROL, SODIUM ION, ... | | Authors: | Gushchin, I, Shevchenko, V, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a light-driven sodium pump.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4XTO
 
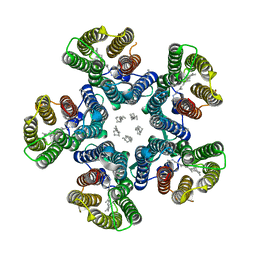 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric red form, pH 5.6 | | Descriptor: | EICOSANE, SODIUM ION, Sodium pumping rhodopsin | | Authors: | Gushchin, I, Shevchenko, V, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a light-driven sodium pump.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4XTN
 
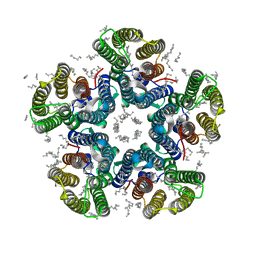 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric red form, pH 4.9 | | Descriptor: | EICOSANE, SODIUM ION, Sodium pumping rhodopsin, ... | | Authors: | Gushchin, I, Shevchenko, V, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a light-driven sodium pump.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7PEN
 
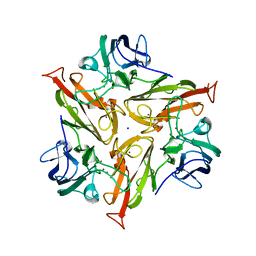 | |
7PES
 
 | | Crystal Structure of Two-Domain Laccase mutant M199G from Streptomyces griseoflavus | | Descriptor: | COPPER (II) ION, OXYGEN MOLECULE, SODIUM ION, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Kolyadenko, I. | | Deposit date: | 2021-08-11 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering the Catalytic Properties of Two-Domain Laccase from Streptomyces griseoflavus Ac-993.
Int J Mol Sci, 23, 2021
|
|
7PUH
 
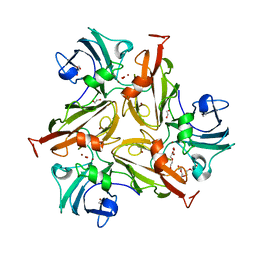 | | Crystal Structure of Two-Domain Laccase mutant H165A/R240H from Streptomyces griseoflavus | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kolyadenko, I, Tishchenko, S, Gabdulkhakov, A. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Engineering the Catalytic Properties of Two-Domain Laccase from Streptomyces griseoflavus Ac-993.
Int J Mol Sci, 23, 2021
|
|
7PTM
 
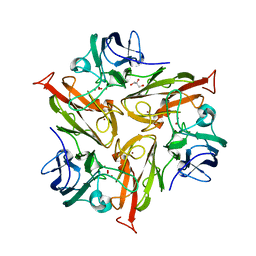 | | Crystal Structure of Two-Domain Laccase mutant M199G/R240H from Streptomyces griseoflavus | | Descriptor: | COPPER (II) ION, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Kolyadenko, I. | | Deposit date: | 2021-09-27 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering the Catalytic Properties of Two-Domain Laccase from Streptomyces griseoflavus Ac-993.
Int J Mol Sci, 23, 2021
|
|
7PU0
 
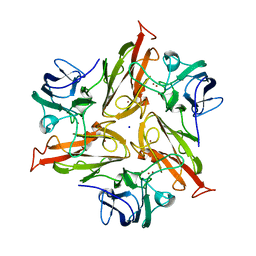 | |
7PFR
 
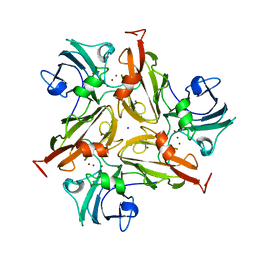 | |
6T0L
 
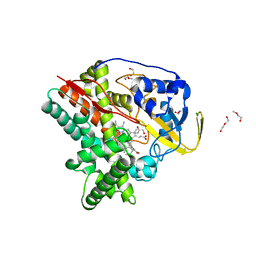 | | Crystal structure of CYP124 in complex with inhibitor compound 5' | | Descriptor: | CHLORIDE ION, CYP124 in complex with inhibitor compound 5', DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
6T0G
 
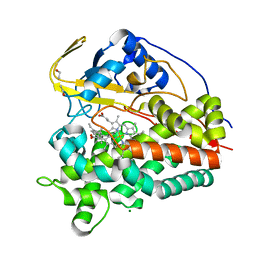 | | Crystal structure of CYP124 in complex with vitamin D3 | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, MAGNESIUM ION, Methyl-branched lipid omega-hydroxylase, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
6T0H
 
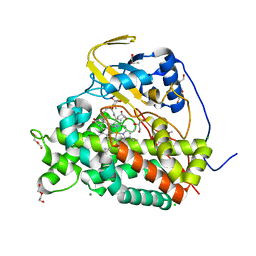 | | Crystal structure of CYP124 in complex with 1-alpha-hydroxy-vitamin D3 | | Descriptor: | 1-alpha-hydroxy-vitamin D3, CHLORIDE ION, CYP124 in complex with inhibitor carbethoxyhexyl imidazole, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
6T0F
 
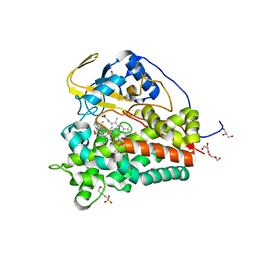 | | Crystal structure of CYP124 in complex with cholest-4-en-3-one | | Descriptor: | (8ALPHA,9BETA)-CHOLEST-4-EN-3-ONE, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
6T0K
 
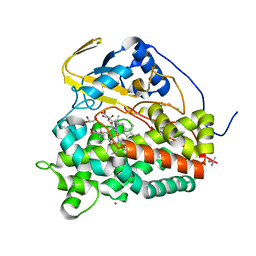 | | Crystal structure of CYP124 in complex with inhibitor carbethoxyhexyl imidazole | | Descriptor: | CHLORIDE ION, CYP124 in complex with inhibitor carbethoxyhexyl imidazole, GLYCEROL, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
8AMP
 
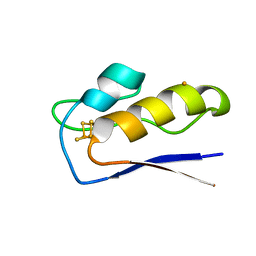 | | Crystal structure of M.tuberculosis ferredoxin Fdx | | Descriptor: | FE (III) ION, FE3-S4 CLUSTER, Possible ferredoxin | | Authors: | Bukhdruker, S, Kavaleuski, A, Marin, E, Kapranov, I, Mishin, A, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-08-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
8AMO
 
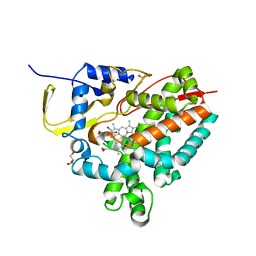 | | Crystal structure of M. tuberculosis CYP143 | | Descriptor: | CHLORIDE ION, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bukhdruker, S, Varaksa, T, Grudo, A, Marin, E, Kapranov, I, Shevtsov, M, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-08-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
7ZB9
 
 | | Crystal structure of CYP124 in complex with inhibitor carbethoxyhexyl imidazole in the absence of glycerol (NoCryo) | | Descriptor: | CHLORIDE ION, CYP124 in complex with inhibitor carbethoxyhexyl imidazole, MAGNESIUM ION, ... | | Authors: | Bukhdruker, S, Varaksa, T, Marin, E, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-03-23 | | Release date: | 2023-01-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into the effects of glycerol on ligand binding to cytochrome P450.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
