6R8N
 
 | | STRUCTURE DETERMINATION OF THE TETRAHEDRAL AMINOPEPTIDASE TET2 FROM P. HORIKOSHII BY USE OF COMBINED SOLID-STATE NMR, SOLUTION-STATE NMR AND EM DATA 4.1 A, FOLLOWED BY REAL_SPACE_REFINEMENT AT 4.1 A | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Colletier, J.-P, Gauto, D, Estrozi, L, Favier, A, Effantin, G, Schoehn, G, Boisbouvier, J, Schanda, P. | | Deposit date: | 2019-04-02 | | Release date: | 2019-08-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
8KCQ
 
 | | Solution structures of the N-terminal divergent caplonin homology (NN-CH) domains of human intraflagellar transport protein 54 | | Descriptor: | TRAF3-interacting protein 1 | | Authors: | Dang, W, Kuwasako, K, He, F, Takahashi, M, Tsuda, K, Nagata, T, Tanaka, A, Kobayashi, N, Kigawa, T, Guentert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2023-08-08 | | Release date: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structure of the N-terminal divergent calponin homology (NN-CH) domain of human intraflagellar transport protein 54.
Biomol.Nmr Assign., 18, 2024
|
|
5ZD0
 
 | | Solution structure of human ubiquitin with three alanine mutations in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | ubiquitin | | Authors: | Tanaka, T, Ikeya, T, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-02-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5Z4B
 
 | | GB1 structure determination in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | Protein LG | | Authors: | Tanaka, T, Teppei, I, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5ZCZ
 
 | | Solution structure of the T. Thermophilus HB8 TTHA1718 protein in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | Heavy metal binding protein | | Authors: | Tanaka, T, Teppei, I, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-02-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
8QAQ
 
 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability. Conformation 1 of omphalotin A in apolar solvents. | | Descriptor: | TRP-MVA-ILE-MVA-MVA-SAR-MVA-IML-SAR-VAL-IML-SAR | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
8Q7J
 
 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability | | Descriptor: | CYCLOSPORIN A | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
8QAS
 
 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability. NMR structure of Omphalotin A in methanol / water indoleOut conformation. | | Descriptor: | TRP-MVA-ILE-MVA-MVA-SAR-MVA-IML-SAR-VAL-IML-SAR | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
8QBP
 
 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability. NMR structure of Omphalotin A in methanol / water indoleOut conformation. | | Descriptor: | TRP-MVA-ILE-MVA-MVA-SAR-MVA-IML-SAR-VAL-IML-SAR | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
6F3K
 
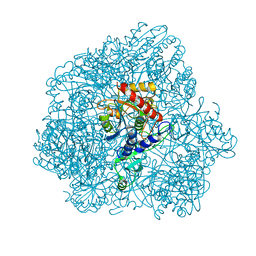 | | Combined solid-state NMR, solution-state NMR and EM data for structure determination of the tetrahedral aminopeptidase TET2 from P. horikoshii | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Gauto, D.F, Estrozi, L.F, Schwieters, C.D, Effantin, G, Macek, P, Sounier, R, Kerfah, R, Sivertsen, A.C, Colletier, J.P, Boisbouvier, J, Schoehn, G, Favier, A, Schanda, P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-03-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLID-STATE NMR, SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
6QK5
 
 | |
6QK6
 
 | |
5N2O
 
 | | Structure Of P63 SAM Domain L514F Mutant Causative Of AEC Syndrome | | Descriptor: | Tumor protein 63 | | Authors: | Rinnenthal, J, Wuerz, J.M, Osterburg, C, Guentert, P, Doetsch, V. | | Deposit date: | 2017-02-08 | | Release date: | 2018-02-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein aggregation of the p63 transcription factor underlies severe skin fragility in AEC syndrome.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7FBR
 
 | |
7FBV
 
 | |
6ESP
 
 | |
5HOB
 
 | | p73 homo-tetramerization domain mutant I | | Descriptor: | MAGNESIUM ION, Tumor protein p73 | | Authors: | Coutandin, D, Krojer, T, Salah, E, Mathea, S, Knapp, S, Dotsch, V. | | Deposit date: | 2016-01-19 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.220013 Å) | | Cite: | Structural basis of p63/p73 hetero-tetramerization
To Be Published
|
|
5HOC
 
 | | p73 homo-tetramerization domain mutant II | | Descriptor: | Tumor protein p73 | | Authors: | Coutandin, D, Krojer, T, Salah, E, Mathea, S, Sumyk, M, Knapp, S, Dotsch, V. | | Deposit date: | 2016-01-19 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.36007786 Å) | | Cite: | Mechanism of TAp73 inhibition by Delta Np63 and structural basis of p63/p73 hetero-tetramerization.
Cell Death Differ., 23, 2016
|
|
6TUB
 
 | | Beta-endorphin amyloid fibril | | Descriptor: | Beta-endorphin | | Authors: | Verasdonck, J, Seuring, C, Gath, J, Ghosh, D, Nespovitaya, N, Waelti, M.A, Maji, S, Cadalbert, R, Boeckmann, A, Guentert, P, Meier, B.H, Riek, R. | | Deposit date: | 2020-01-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The three-dimensional structure of human beta-endorphin amyloid fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5LAO
 
 | | S-nitrosylated 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5LAM
 
 | | Refined 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
1UKX
 
 | | Solution structure of the RWD domain of mouse GCN2 | | Descriptor: | GCN2 eIF2alpha kinase | | Authors: | Nameki, N, Yoneyama, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RWD domain of the mouse GCN2 protein.
Protein Sci., 13, 2004
|
|
1J26
 
 | | Solution structure of a putative peptidyl-tRNA hydrolase domain in a mouse hypothetical protein | | Descriptor: | immature colon carcinoma transcript 1 | | Authors: | Nameki, N, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-25 | | Release date: | 2004-06-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of the mitochondrial protein ICT1 that is essential for cell vitality
J.Mol.Biol., 2010
|
|
1N27
 
 | | Solution structure of the PWWP domain of mouse Hepatoma-derived growth factor, related protein 3 | | Descriptor: | Hepatoma-derived growth factor, related protein 3 | | Authors: | Nameki, N, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-22 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PWWP domain of the hepatoma-derived growth factor family.
Protein Sci., 14, 2005
|
|
2YUE
 
 | | Solution structure of the NEUZ (NHR) domain in Neuralized from Drosophila melanogaster | | Descriptor: | Protein neuralized | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of the NHR1 domain of the Drosophila neuralized E3 ligase in the notch signaling pathway.
J.Mol.Biol., 393, 2009
|
|
