1AA2
 
 | |
5H55
 
 | | Mdm12 from K. lactis | | Descriptor: | Mitochondrial distribution and morphology protein 12 | | Authors: | Kawano, S, Quinbara, S. | | Deposit date: | 2016-11-04 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES
J. Cell Biol., 217, 2018
|
|
4CVH
 
 | | Crystal structure of human isoprenoid synthase domain-containing protein | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ISOPRENOID SYNTHASE DOMAIN-CONTAINING PROTEIN, ... | | Authors: | Kopec, J, Froese, D.S, Krojer, T, Newman, J, Kiyani, W, Goubin, S, Strain-Damerell, C, Vollmar, M, von Delft, F, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Lefeber, D.J, Yue, W.W. | | Deposit date: | 2014-03-27 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Human Ispd is a Cytidyltransferase Required for Dystroglycan O-Mannosylation.
Chem.Biol., 22, 2015
|
|
9BFB
 
 | | Crystal structure of BRAF kinase domain with PF-07284890 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-{2-chloro-3-[(3,5-dimethyl-4-oxo-3,4-dihydroquinazolin-6-yl)amino]-4-fluorophenyl}-3-fluoropropane-1-sulfonamide, ... | | Authors: | Mou, T.-C. | | Deposit date: | 2024-04-17 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification of the Clinical Candidate PF-07284890 ( ARRY-461 ), a Highly Potent and Brain Penetrant BRAF Inhibitor for the Treatment of Cancer.
J.Med.Chem., 67, 2024
|
|
5Z5R
 
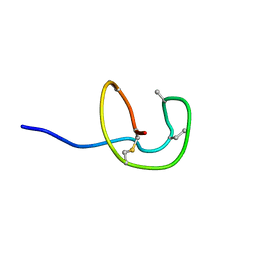 | | Nukacin ISK-1 in inactive state | | Descriptor: | Lantibiotic nukacin | | Authors: | Kohda, D, Fujinami, D. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | The lantibiotic nukacin ISK-1 exists in an equilibrium between active and inactive lipid-II binding states.
Commun Biol, 1, 2018
|
|
6S4I
 
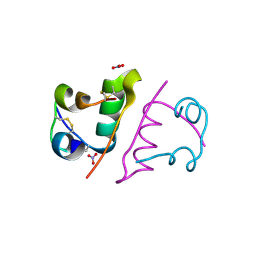 | |
6S4J
 
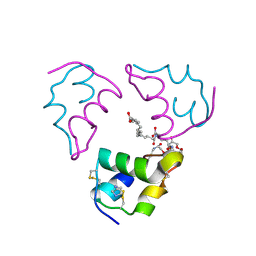 | |
2MP8
 
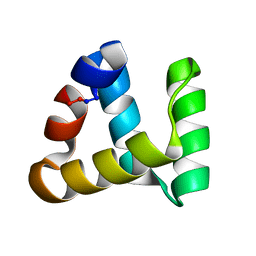 | | NMR structure of NKR-5-3B | | Descriptor: | NKR-5-3B | | Authors: | Rosengren, K.J, Craik, D.J. | | Deposit date: | 2014-05-13 | | Release date: | 2015-05-13 | | Last modified: | 2016-06-01 | | Method: | SOLUTION NMR | | Cite: | Identification, Characterization, and Three-Dimensional Structure of the Novel Circular Bacteriocin, Enterocin NKR-5-3B, from Enterococcus faecium
Biochemistry, 54, 2015
|
|
6S34
 
 | |
8BS0
 
 | | Room-temperature structure of Pedobacter heparinus N-acetylglucosamine 2-epimerase at 80 MPa helium gas pressure in a sapphire capillary | | Descriptor: | CHLORIDE ION, N-acylglucosamine 2-epimerase, PHOSPHATE ION | | Authors: | Lieske, J, Saouane, S, Assmann, M, Zaun, H, Kuballa, J, Meents, A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
8BRZ
 
 | | Room-temperature structure of Pedobacter heparinus N-acetylglucosamine 2-epimerase at 52 MPa helium gas pressure in a sapphire capillary | | Descriptor: | CHLORIDE ION, N-acylglucosamine 2-epimerase, PHOSPHATE ION | | Authors: | Lieske, J, Saouane, S, Assmann, M, Zaun, H, Kuballa, J, Meents, A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
8BRY
 
 | | Room-temperature structure of Pedobacter heparinus N-acetylglucosamine 2-epimerase at atmospheric pressure | | Descriptor: | CHLORIDE ION, N-acylglucosamine 2-epimerase, PHOSPHATE ION | | Authors: | Lieske, J, Saouane, S, Assmann, M, Zaun, H, Kuballa, J, Meents, A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
8BS1
 
 | | Room-temperature structure of SARS-CoV-2 Main protease at atmospheric pressure | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Lieske, J, Saouane, S, Guenther, S, Reinke, P.Y.A, Rahmani Mashhour, A, Meents, A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
8BS2
 
 | | Room-temperature structure of SARS-CoV-2 Main protease at 104 MPa helium gas pressure in a sapphire capillary | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Lieske, J, Saouane, S, Guenther, S, Reinke, P.Y.A, Rahmani Mashhour, A, Meents, A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
5Z5Q
 
 | | Nukacin ISK-1 in active state | | Descriptor: | Lantibiotic nukacin | | Authors: | Kohda, D, Fujinami, D. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | The lantibiotic nukacin ISK-1 exists in an equilibrium between active and inactive lipid-II binding states.
Commun Biol, 1, 2018
|
|
6U2G
 
 | | BRAF-MEK complex with AMP-PCP bound to BRAF | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Liau, N.P.D, Wendorff, T, Hymowitz, S, Sudhamsu, J. | | Deposit date: | 2019-08-19 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | Negative regulation of RAF kinase activity by ATP is overcome by 14-3-3-induced dimerization.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6TG6
 
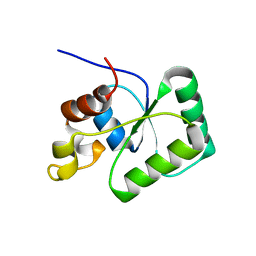 | | Toprim domain of RNase M5 | | Descriptor: | Ribonuclease M5 | | Authors: | Oerum, S, Catala, M, Tisne, C. | | Deposit date: | 2019-11-15 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
8Q85
 
 | |
8Q84
 
 | |
8QAU
 
 | | Outer kinetochore Ndc80-Dam1 alpha/beta-tubulin complex | | Descriptor: | DASH complex subunit DAM1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Muir, K.W, Barford, D. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Mechanism of outer kinetochore assembly on microtubules and its regulation by mitotic error correction
Biorxiv, 2023
|
|
7Q79
 
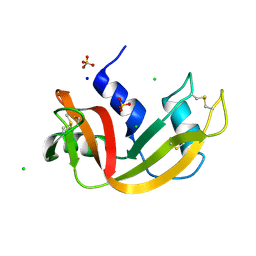 | | Room temperature structure of RNase A at 100 MPa helium gas pressure in a sapphire capillary | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SODIUM ION, ... | | Authors: | Lieske, J, Guenther, S, Saouane, S, Meents, A. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fixed-target high-pressure macromolecular crystallography
To Be Published
|
|
7Q75
 
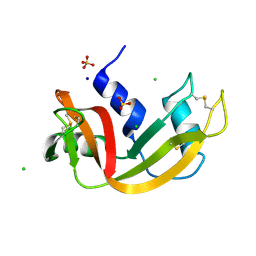 | | Room temperature structure of RNase A at atmospheric pressure | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SODIUM ION, ... | | Authors: | Lieske, J, Guenther, S, Saouane, S, Meents, A. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Fixed-target high-pressure macromolecular crystallography
To Be Published
|
|
7Q7F
 
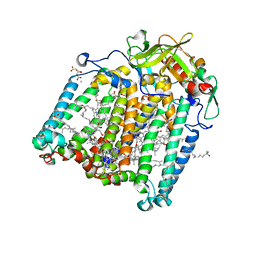 | | Room temperature structure of the Rhodobacter Sphaeroides Photosynthetic Reaction Center F(M197)H mutant at atmospheric pressure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Lieske, J, Guenther, S, Saouane, S, Selikhanov, G.K, Gabdulkhakov, A.G, Meents, A. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Fixed-target high-pressure macromolecular crystallography
To Be Published
|
|
7Q78
 
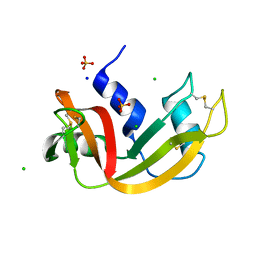 | | Room temperature structure of RNase A at 72 MPa helium gas pressure in a sapphire capillary | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SODIUM ION, ... | | Authors: | Lieske, J, Guenther, S, Saouane, S, Meents, A. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Fixed-target high-pressure macromolecular crystallography
To Be Published
|
|
7Q7N
 
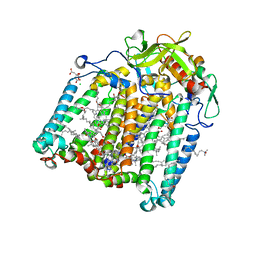 | | Room temperature structure of the Rhodobacter Sphaeroides Photosynthetic Reaction Center F(M197)H mutant at 120 MPa helium gas pressure in a sapphire capillary | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Lieske, J, Guenther, S, Saouane, S, Selikhanov, G.K, Gabdulkhakov, A.G, Meents, A. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Fixed-target high-pressure macromolecular crystallography
To Be Published
|
|
