7UR5
 
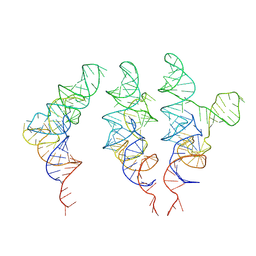 | | allo-tRNAUTu1 in the A, P, and E sites of the E. coli ribosome | | Descriptor: | allo-tRNAUTu1 | | Authors: | Zhang, J, Krahn, N, Prabhakar, A, Vargas-Rodriguez, O, Krupkin, M, Fu, Z, Acosta-Reyes, F.J, Ge, X, Choi, J, Crnkovic, A, Ehrenberg, M, Viani Puglisi, E, Soll, D, Puglisi, J. | | Deposit date: | 2022-04-21 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Uncovering translation roadblocks during the development of a synthetic tRNA.
Nucleic Acids Res., 50, 2022
|
|
7URM
 
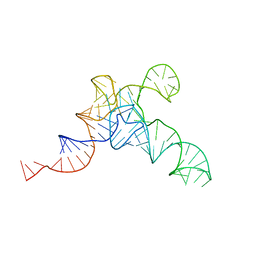 | | allo-tRNAUTu1A in the P site of the E. coli ribosome | | Descriptor: | allo-tRNAUTU1A | | Authors: | Zhang, J, Prabhakar, A, Krahn, N, Vargas-Rodriguez, O, Krupkin, M, Fu, Z, Acosta-Reyes, F.J, Ge, X, Choi, J, Crnkovic, A, Ehrenberg, M, Viani Puglisi, E, Soll, D, Puglisi, J. | | Deposit date: | 2022-04-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Uncovering translation roadblocks during the development of a synthetic tRNA.
Nucleic Acids Res., 50, 2022
|
|
2FAP
 
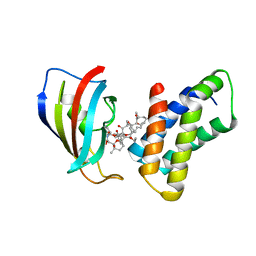 | | THE STRUCTURE OF THE IMMUNOPHILIN-IMMUNOSUPPRESSANT FKBP12-(C16)-ETHOXY RAPAMYCIN COMPLEX INTERACTING WITH HUMA | | Descriptor: | C49-METHYL RAPAMYCIN, FK506-BINDING PROTEIN, FRAP | | Authors: | Liang, J, Choi, J, Clardy, J. | | Deposit date: | 1998-09-22 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined structure of the FKBP12-rapamycin-FRB ternary complex at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5YPU
 
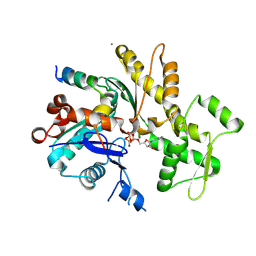 | | Crystal structure of an actin monomer in complex with the nucleator Cordon-Bleu MET72NLE WH2-motif peptide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Scipion, C.P.M, Wongsantichon, J, Ferrer, F.J, Yuen, T.Y, Robinson, R.C. | | Deposit date: | 2017-11-03 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for the roles of divalent cations in actin polymerization and activation of ATP hydrolysis
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YVN
 
 | | Human Glutathione Transferase Omega1 | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione S-transferase omega-1, ... | | Authors: | Saisawang, C, Ketterman, A, Wongsantichon, J. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Glutathione transferase Omega 1-1 (GSTO1-1) modulates Akt and MEK1/2 signaling in human neuroblastoma cell SH-SY5Y.
Proteins, 87, 2019
|
|
5YVO
 
 | | Human Glutathione Transferase Omega1 covalently bound to ML175 inhibitor | | Descriptor: | ACETATE ION, Glutathione S-transferase omega-1, N-{3-[(2-chloro-acetyl)-(4-nitro-phenyl)-amino]-propyl}-2,2,2-trifluoro-acetamide, ... | | Authors: | Saisawang, C, Ketterman, A, Wongsantichon, J. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-28 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glutathione transferase Omega 1-1 (GSTO1-1) modulates Akt and MEK1/2 signaling in human neuroblastoma cell SH-SY5Y.
Proteins, 87, 2019
|
|
7ZLK
 
 | | AMC009 SOSIPv5.2 in complex with Fabs ACS101 and ACS124 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACS114 heavy chain, ... | | Authors: | van Schooten, J, Ozorowski, G, Ward, A. | | Deposit date: | 2022-04-15 | | Release date: | 2022-09-21 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
Plos Pathog., 18, 2022
|
|
5K2R
 
 | | Crystal structure of lysozyme | | Descriptor: | Lysozyme C | | Authors: | Ko, S, Choe, J. | | Deposit date: | 2016-05-19 | | Release date: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of lysozyme with nano particles
To Be Published
|
|
5K2N
 
 | |
5K2P
 
 | | Crystal structure of lysozyme | | Descriptor: | Lysozyme C | | Authors: | Ko, S, Choe, J. | | Deposit date: | 2016-05-19 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of lysozyme
To Be Published
|
|
5IS1
 
 | |
5K2S
 
 | | Lysozyme with nano particles | | Descriptor: | Lysozyme C | | Authors: | Ko, S, Choe, J. | | Deposit date: | 2016-05-19 | | Release date: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of lysozyme with nano particles
To Be Published
|
|
5K2K
 
 | | Crystal structure of lysozyme | | Descriptor: | Lysozyme C | | Authors: | Ko, S, Choe, J. | | Deposit date: | 2016-05-19 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of lysozyme
To Be Published
|
|
5K2Q
 
 | | Lysozyme with nano particles | | Descriptor: | Lysozyme C | | Authors: | Ko, S, Choe, J. | | Deposit date: | 2016-05-19 | | Release date: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of lysozyme with nano particle
To Be Published
|
|
5LJP
 
 | |
6TPK
 
 | | Crystal structure of the human oxytocin receptor | | Descriptor: | (3~{R},6~{R})-6-[(2~{S})-butan-2-yl]-3-(2,3-dihydro-1~{H}-inden-2-yl)-1-[(1~{R})-1-(2-methyl-1,3-oxazol-4-yl)-2-morpholin-4-yl-2-oxidanylidene-ethyl]piperazine-2,5-dione, CHOLESTEROL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Waltenspuehl, Y, Schoeppe, J, Ehrenmann, J, Kummer, L, Plueckthun, A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-08-05 | | Last modified: | 2020-09-02 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the human oxytocin receptor.
Sci Adv, 6, 2020
|
|
1TXM
 
 | | SCORPION TOXIN (MAUROTOXIN) FROM SCORPIO MAURUS, NMR, 35 STRUCTURES | | Descriptor: | MAUROTOXIN | | Authors: | Blanc, E, Sabatier, J.-M, Kharrat, R, Meunier, S, El Ayeb, M, Van Rietschoten, J, Darbon, H. | | Deposit date: | 1996-11-19 | | Release date: | 1997-06-05 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of maurotoxin, a scorpion toxin from Scorpio maurus, with high affinity for voltage-gated potassium channels.
Proteins, 29, 1997
|
|
5GJ7
 
 | | putative Acyl-CoA dehydrogenase | | Descriptor: | Acyl-CoA dehydrogenase type 2 domain protein | | Authors: | Moon, H, Choe, J. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Putative Acyl CoA dehydrogenase
To Be Published
|
|
5GUY
 
 | | Structural and functional study of chuY from E. coli CFT073 strain | | Descriptor: | Uncharacterized protein chuY | | Authors: | Kim, H, Choi, J, HA, S.C, Chaurasia, A.K, Kim, D, Kim, K.K. | | Deposit date: | 2016-08-31 | | Release date: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural and functional study of ChuY from Escherichia coli strain CFT073
Biochem. Biophys. Res. Commun., 482, 2017
|
|
6AVV
 
 | |
6AVY
 
 | |
6ATX
 
 | |
6AZC
 
 | |
6AVW
 
 | |
2PUW
 
 | | The crystal structure of isomerase domain of glucosamine-6-phosphate synthase from Candida albicans | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, CHLORIDE ION, isomerase domain of glutamine-fructose-6-phosphate transaminase (isomerizing) | | Authors: | Raczynska, J, Olchowy, J, Milewski, S, Rypniewski, W. | | Deposit date: | 2007-05-09 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.151 Å) | | Cite: | The Crystal and Solution Studies of Glucosamine-6-phosphate Synthase from Candida albicans
J.Mol.Biol., 372, 2007
|
|
