4U1O
 
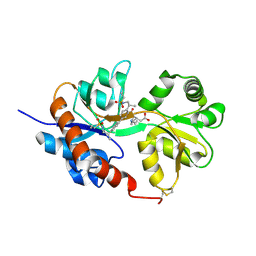 | | GluA2flip sLBD complexed with kainate and (R,R)-2b crystal form C | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-15 | | Release date: | 2014-08-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8501 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
9U9B
 
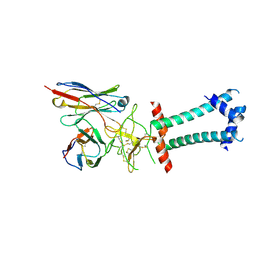 | | HBsAg in complex with HBC34 Fab | | Descriptor: | HBC34 Fab Heavy Chain, HBC34 Fab Light Chain, Middle S protein | | Authors: | Chen, L, He, X. | | Deposit date: | 2025-03-27 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural polymorphism of the antigenic loop in HBV surface antigen dictates binding of diverse neutralizing antibodies.
Cell Discov, 11, 2025
|
|
9IYY
 
 | | Structure of HBsAg in complex with Fab H006 | | Descriptor: | Large envelope protein, heavy chain of antibody H006 Fab, light chain of antibody H006 Fab | | Authors: | Chen, L, He, X. | | Deposit date: | 2024-07-31 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural polymorphism of the antigenic loop in HBV surface antigen dictates binding of diverse neutralizing antibodies.
Cell Discov, 11, 2025
|
|
9JT1
 
 | | Structure of HBsAg in complex with FabHBC and FabGC1102 | | Descriptor: | Large envelope protein, heavy chain of GC1102, heavy chain of HBC, ... | | Authors: | Chen, L, He, X, Tao, W. | | Deposit date: | 2024-10-01 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural polymorphism of the antigenic loop in HBV surface antigen dictates binding of diverse neutralizing antibodies.
Cell Discov, 11, 2025
|
|
6L48
 
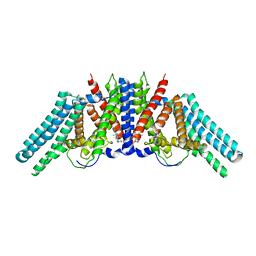 | |
6L47
 
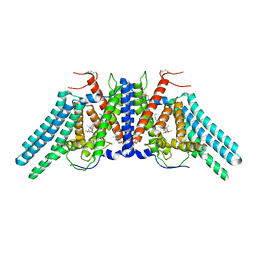 | | Structure of the human sterol O-acyltransferase 1 in complex with CI-976 | | Descriptor: | 2,2-dimethyl-N-(2,4,6-trimethoxyphenyl)dodecanamide, CHOLESTEROL, Sterol O-acyltransferase 1 | | Authors: | Chen, L, Guan, C, Niu, Y. | | Deposit date: | 2019-10-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor.
Nat Commun, 11, 2020
|
|
3BWH
 
 | | Atomic resolution structure of cucurmosin, a novel type 1 RIP from the sarcocarp of Cucurbita moschata | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, beta-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-3)][alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L. | | Deposit date: | 2008-01-09 | | Release date: | 2008-10-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution structure of cucurmosin, a novel type 1 ribosome-inactivating protein from the sarcocarp of Cucurbita moschata.
J.Struct.Biol., 164, 2008
|
|
3DAE
 
 | |
5F9W
 
 | | Crystal structure of broadly neutralizing VH1-46 germline-derived CD4-binding site-directed antibody CH235 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of CH235-lineage antibody CH235, Light chain of CH235-lineage antibody CH235, ... | | Authors: | Chen, L, Zhou, T, Kwong, P.D. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.8911 Å) | | Cite: | Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody.
Cell, 165, 2016
|
|
3WMC
 
 | | Crystal structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with naphthalimide derivative Q2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(dimethylamino)-2-(2-{[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]amino}ethyl)-1H-benzo[de]isoquinoline-1,3(2H)-dione, Beta-hexosaminidase | | Authors: | Chen, L, Zhou, Y, Chen, L, Yang, Q. | | Deposit date: | 2013-11-16 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | A crystal structure-guided rational design switching non-carbohydrate inhibitors' specificity between two beta-GlcNAcase homologs
Sci Rep, 4, 2014
|
|
1AAN
 
 | | CRYSTAL STRUCTURE ANALYSIS OF AMICYANIN AND APOAMICYANIN FROM PARACOCCUS DENITRIFICANS AT 2.0 ANGSTROMS AND 1.8 ANGSTROMS RESOLUTION | | Descriptor: | AMICYANIN, COPPER (II) ION | | Authors: | Chen, L, Durley, R.C.E, Lim, L.W, Mathews, F.S. | | Deposit date: | 1992-04-09 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure analysis of amicyanin and apoamicyanin from Paracoccus denitrificans at 2.0 A and 1.8 A resolution.
Protein Sci., 2, 1993
|
|
6WC5
 
 | | Crystal Structure of a Ternary MEF2B/NKX2-5/myocardin enhancer DNA Complex | | Descriptor: | Homeobox protein Nkx-2.5, Myocardin enhancer DNA, Myocyte-specific enhancer factor 2B | | Authors: | Chen, L, Lei, X. | | Deposit date: | 2020-03-29 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Ternary Complexes of MEF2 and NKX2-5 Bound to DNA Reveal a Disease Related Protein-Protein Interaction Interface.
J.Mol.Biol., 432, 2020
|
|
1RHP
 
 | |
1TQE
 
 | | Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2 | | Descriptor: | Histone deacetylase 9, MEF2 binding site of nur77 promoter, Myocyte-specific enhancer factor 2B | | Authors: | Chen, L, Han, A, He, J, Wu, Y, Liu, J.O. | | Deposit date: | 2004-06-17 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of Recruitment of Class II Histone Deacetylases by Myocyte Enhancer Factor-2.
J.Mol.Biol., 345, 2005
|
|
2LOZ
 
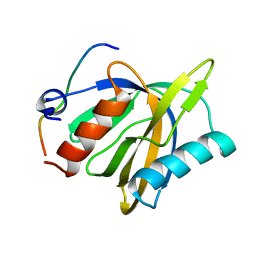 | | The novel binding mode of DLC1 and Tensin2 PTB domain | | Descriptor: | Rho GTPase-activating protein 7, Tensin-like C1 domain-containing phosphatase | | Authors: | Liu, C, Chen, L, Zhu, G. | | Deposit date: | 2012-01-29 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the phosphotyrosine binding (PTB) domain of human tensin2 protein in complex with deleted in liver cancer 1 (DLC1) peptide reveals a novel peptide binding mode.
J.Biol.Chem., 287, 2012
|
|
9IYX
 
 | |
9EK1
 
 | | HIV-1 mature WT matrix protein p17 lattice | | Descriptor: | MYRISTIC ACID, Matrix protein p17 | | Authors: | Rey, J.S, Perilla, J.R, Chen, L, Zhang, P. | | Deposit date: | 2024-11-30 | | Release date: | 2025-01-08 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structural maturation of the matrix lattice is not required for HIV-1 particle infectivity.
Sci Adv, 11, 2025
|
|
9EK3
 
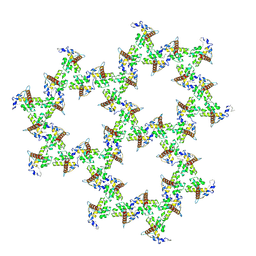 | | HIV-1 immature WT matrix protein p17 lattice | | Descriptor: | MYRISTIC ACID, Matrix protein p17 | | Authors: | Rey, J.S, Perilla, J.R, Chen, L, Zhang, P. | | Deposit date: | 2024-11-30 | | Release date: | 2025-01-08 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural maturation of the matrix lattice is not required for HIV-1 particle infectivity.
Sci Adv, 11, 2025
|
|
9EK2
 
 | | HIV-1 immature L20K/E73K/A82T matrix protein p17 lattice | | Descriptor: | MYRISTIC ACID, Matrix protein p17 | | Authors: | Rey, J.S, Perilla, J.R, Chen, L, Zhang, P. | | Deposit date: | 2024-11-30 | | Release date: | 2025-01-08 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Structural maturation of the matrix lattice is not required for HIV-1 particle infectivity.
Sci Adv, 11, 2025
|
|
4U1W
 
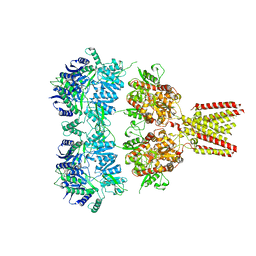 | | Full length GluA2-kainate-(R,R)-2b complex crystal form A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U1X
 
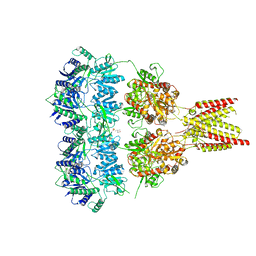 | | Full length GluA2-kainate-(R,R)-2b complex crystal form B | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
2HJD
 
 | |
5TDG
 
 | |
5TDL
 
 | |
3HI1
 
 | | Structure of HIV-1 gp120 (core with V3) in Complex with CD4-Binding-Site Antibody F105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F105 Heavy Chain, F105 Light Chain, ... | | Authors: | Kwon, Y.D, Chen, L, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z, Zhang, M.-Y, Arthos, J, Burton, D.R, Dimitrov, D, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-05-18 | | Release date: | 2009-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
