5VR4
 
 | | RNA octamer containing 2'-F-4'-OMe U. | | Descriptor: | COBALT TETRAAMMINE ION, RNA (5'-R(*CP*GP*AP*AP*(UMO)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2017-05-10 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4'-C-Methoxy-2'-deoxy-2'-fluoro Modified Ribonucleotides Improve Metabolic Stability and Elicit Efficient RNAi-Mediated Gene Silencing.
J. Am. Chem. Soc., 139, 2017
|
|
5DER
 
 | | RNA oligonucleotide containing (R)-C5'-ME-2'F U | | Descriptor: | RNA oligonucleotide containing (R)-C5'-Me-2'-FU | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2015-08-25 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Duplex Thermodynamic Stability and Enhanced Nuclease Resistance of 5'-C-Methyl Pyrimidine-Modified Oligonucleotides.
J.Org.Chem., 81, 2016
|
|
5DEK
 
 | | RNA octamer containing dT | | Descriptor: | COBALT HEXAMMINE(III), RNA oligonucleotide containing dT | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2015-08-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | Structural Basis of Duplex Thermodynamic Stability and Enhanced Nuclease Resistance of 5'-C-Methyl Pyrimidine-Modified Oligonucleotides.
J.Org.Chem., 81, 2016
|
|
5D8T
 
 | | RNA octamer containing (S)-5' methyl, 2'-F U. | | Descriptor: | COBALT HEXAMMINE(III), RNA oligonucleotide containing (S)-C5'-Me-2'-FU | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2015-08-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of Duplex Thermodynamic Stability and Enhanced Nuclease Resistance of 5'-C-Methyl Pyrimidine-Modified Oligonucleotides.
J.Org.Chem., 81, 2016
|
|
5FN1
 
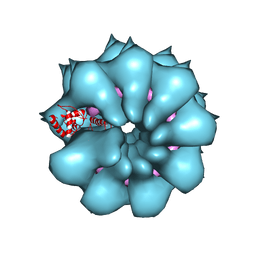 | | Electron cryo-microscopy of filamentous flexible virus PepMV (Pepino Mosaic Virus) | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP)-3', COAT PROTEIN | | Authors: | Agirrezabala, X, Mendez-Lopez, E, Lasso, G, Sanchez-Pina, M.A, Aranda, M.A, Valle, M. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The near-atomic cryoEM structure of a flexible filamentous plant virus shows homology of its coat protein with nucleoproteins of animal viruses.
Elife, 4, 2015
|
|
5ODV
 
 | | Structure of Watermelon mosaic virus potyvirus. | | Descriptor: | RNA (5'-R(P*UP*UP*UP*UP*U)-3'), coat protein | | Authors: | Zamora, M, Mendez-Lopez, E, Agirrezabala, X, Cuesta, R, Lavin, J.L, Sanchez-Pina, M.A, Aranda, M, Valle, M. | | Deposit date: | 2017-07-06 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Potyvirus virion structure shows conserved protein fold and RNA binding site in ssRNA viruses.
Sci Adv, 3, 2017
|
|
3KW6
 
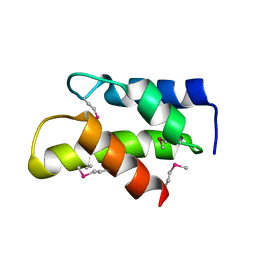 | | Crystal Structure of a domain of 26S proteasome regulatory subunit 8 from homo sapiens. Northeast Structural Genomics Consortium target id HR3102A | | Descriptor: | 26S protease regulatory subunit 8 | | Authors: | Seetharaman, J, Su, M, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-22 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a domain of 26S proteasome regulatory subunit 8 from homo sapiens. Northeast Structural Genomics Consortium target id HR3102A
To be Published
|
|
5K4Z
 
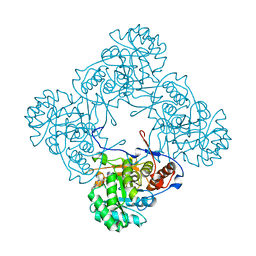 | | M. thermoresistible IMPDH in complex with IMP and Compound 6 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ~{N}-(4-fluorophenyl)-4-(2~{H}-indazol-6-ylsulfamoyl)-3,5-dimethyl-1~{H}-pyrrole-2-carboxamide | | Authors: | Pacitto, A, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-05-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Essential but Not Vulnerable: Indazole Sulfonamides Targeting Inosine Monophosphate Dehydrogenase as Potential Leads against Mycobacterium tuberculosis.
ACS Infect Dis, 3, 2017
|
|
8OKH
 
 | |
5K4X
 
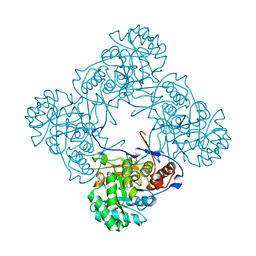 | | M. thermoresistible IMPDH in complex with IMP and Compound 1 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ~{N}-(2~{H}-indazol-6-yl)-3,5-dimethyl-1~{H}-pyrazole-4-sulfonamide | | Authors: | Pacitto, A, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-05-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Essential but Not Vulnerable: Indazole Sulfonamides Targeting Inosine Monophosphate Dehydrogenase as Potential Leads against Mycobacterium tuberculosis.
ACS Infect Dis, 3, 2017
|
|
3G02
 
 | |
2HC5
 
 | | Solution NMR Structure of Protein yvyC from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR482. | | Descriptor: | Hypothetical protein yvyC | | Authors: | Eletsky, A, Liu, G, Atreya, H.S, Sukumaran, D.K, Wang, D, Cunningham, K, Janjua, H, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-15 | | Release date: | 2006-08-15 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein YvyC from Bacillus subtilis reveals unexpected structural similarity between two PFAM families.
Proteins, 76, 2009
|
|
3G0I
 
 | |
2NWT
 
 | | NMR Structure of Protein UPF0165 protein AF_2212 from Archaeoglobus Fulgidus; Northeast Structural Genomics Consortium Target GR83 | | Descriptor: | UPF0165 protein AF_2212 | | Authors: | Singarapu, K.K, Sukumaran, D.K, Parish, D, Atreya, H.S, Liu, G, Eletsky, A, Chen, C.X, Jiang, M, Cunningham, K, Xiao, R, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-16 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Protein Y2212_ARCFU from Archaeoglobus Fulgidus; Northeast Structural Genomics Consortium Target GR83
To be Published
|
|
9BF2
 
 | | MID domain of Ago2 bound to UMP | | Descriptor: | Protein argonaute-2, URIDINE-5'-MONOPHOSPHATE | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure and Stability of Ago2 MID-Nucleotide Complexes: All-in-One (Drop) His 6 -SUMO Tag Removal, Nucleotide Binding, and Crystal Growth.
Curr Protoc, 4, 2024
|
|
9BF0
 
 | | MID domain of human Argo2 bound to UTP | | Descriptor: | Protein argonaute-2, URIDINE 5'-TRIPHOSPHATE | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure and Stability of Ago2 MID-Nucleotide Complexes: All-in-One (Drop) His 6 -SUMO Tag Removal, Nucleotide Binding, and Crystal Growth.
Curr Protoc, 4, 2024
|
|
9BEZ
 
 | | MID domain of human Argo2 bound to RNA | | Descriptor: | Protein argonaute-2, [(3~{S},4~{R},5~{R})-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-4-oxidanyl-oxolan-3-yl] [oxidanyl(phosphonooxy)phosphoryl] hydrogen phosphate | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Stability of Ago2 MID-Nucleotide Complexes: All-in-One (Drop) His 6 -SUMO Tag Removal, Nucleotide Binding, and Crystal Growth.
Curr Protoc, 4, 2024
|
|
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
5GVP
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and GS654 | | Descriptor: | 3-[3-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-(trifluoromethyl)phenyl]phenyl]propanoic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G. | | Deposit date: | 2016-09-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Antimalarial Inhibitors Targeting Serine Hydroxymethyltransferase (SHMT) with in Vivo Efficacy and Analysis of their Binding Mode Based on X-ray Cocrystal Structures
J. Med. Chem., 60, 2017
|
|
5GVN
 
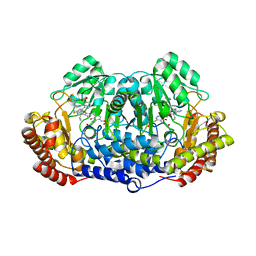 | | Plasmodium vivax SHMT bound with PLP-glycine and GS653 | | Descriptor: | 3-[3-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-fluoranyl-phenyl]phenyl]propanoic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G. | | Deposit date: | 2016-09-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antimalarial Inhibitors Targeting Serine Hydroxymethyltransferase (SHMT) with in Vivo Efficacy and Analysis of their Binding Mode Based on X-ray Cocrystal Structures
J. Med. Chem., 60, 2017
|
|
5GVL
 
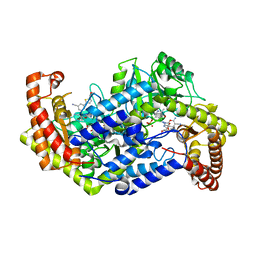 | | Plasmodium vivax SHMT bound with PLP-glycine and GS182 | | Descriptor: | (4~{S})-6-azanyl-4-[3-cyano-5-[5-(methoxymethyl)thiophen-2-yl]phenyl]-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazole-5-carbonitrile, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G. | | Deposit date: | 2016-09-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antimalarial Inhibitors Targeting Serine Hydroxymethyltransferase (SHMT) with in Vivo Efficacy and Analysis of their Binding Mode Based on X-ray Cocrystal Structures
J. Med. Chem., 60, 2017
|
|
5GVM
 
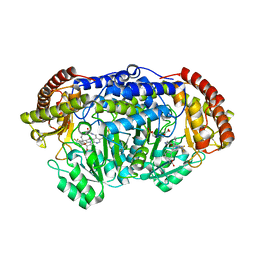 | | Plasmodium vivax SHMT bound with PLP-glycine and GS557 | | Descriptor: | 2-[3-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-(trifluoromethyl)phenyl]phenyl]ethanoic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G. | | Deposit date: | 2016-09-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Antimalarial Inhibitors Targeting Serine Hydroxymethyltransferase (SHMT) with in Vivo Efficacy and Analysis of their Binding Mode Based on X-ray Cocrystal Structures
J. Med. Chem., 60, 2017
|
|
6CVZ
 
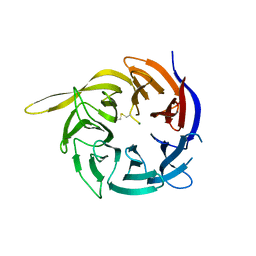 | | Crystal structure of the WD40-repeat of RFWD3 | | Descriptor: | E3 ubiquitin-protein ligase RFWD3, MAGNESIUM ION | | Authors: | DONG, A, LOPPNAU, P, SEITOVA, A, HUTCHINSON, A, TEMPEL, W, WEI, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Target highlights in CASP13: Experimental target structures through the eyes of their authors.
Proteins, 87, 2019
|
|
5GVK
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and GS256 | | Descriptor: | 5-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-cyano-phenyl]-~{N},~{N}-dimethyl-thiophene-2-sulfonamide, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G. | | Deposit date: | 2016-09-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Antimalarial Inhibitors Targeting Serine Hydroxymethyltransferase (SHMT) with in Vivo Efficacy and Analysis of their Binding Mode Based on X-ray Cocrystal Structures
J. Med. Chem., 60, 2017
|
|
6SD8
 
 | | Bd2924 apo-form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable acyl-CoA dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2019-07-26 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Target highlights in CASP13: Experimental target structures through the eyes of their authors.
Proteins, 87, 2019
|
|
