1Y18
 
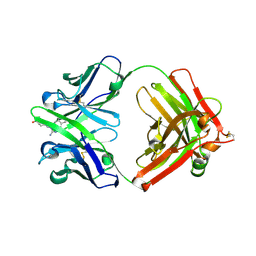 | | Fab fragment of catalytic elimination antibody 34E4 E(H50)D mutant in complex with hapten | | Descriptor: | 2-AMINO-5,6-DIMETHYL-BENZIMIDAZOLE-1-PENTANOIC ACID, CHLORIDE ION, Catalytic antibody 34E4 heavy chain, ... | | Authors: | Debler, E.W, Ito, S, Heine, A, Wilson, I.A. | | Deposit date: | 2004-11-17 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural origins of efficient proton abstraction from carbon by a catalytic antibody
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1Y0L
 
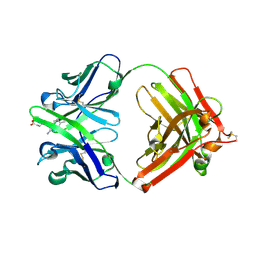 | | Catalytic elimination antibody 34E4 in complex with hapten | | Descriptor: | 2-AMINO-5,6-DIMETHYL-BENZIMIDAZOLE-1-PENTANOIC ACID, CHLORIDE ION, Catalytic Antibody Fab 34E4 Heavy chain, ... | | Authors: | Debler, E.W, Ito, S, Heine, A, Wilson, I.A. | | Deposit date: | 2004-11-15 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural origins of efficient proton abstraction from carbon by a catalytic antibody
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
7LGM
 
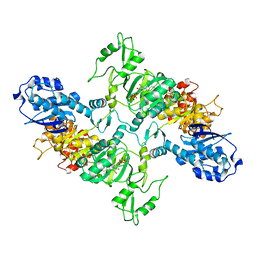 | | Cyanophycin synthetase from A. baylyi DSM587 with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyanophycin synthase | | Authors: | Sharon, I, Haque, A.S, Lahiri, I, Leschziner, A, Schmeing, T.M. | | Deposit date: | 2021-01-20 | | Release date: | 2021-08-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structures and function of the amino acid polymerase cyanophycin synthetase.
Nat.Chem.Biol., 17, 2021
|
|
7LG5
 
 | |
7LGN
 
 | |
3NXF
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | Descriptor: | Retro-Aldolase, SULFATE ION | | Authors: | Althoff, E.A, Jiang, L, Wang, L, Lassila, J.K, Moody, J, Bolduc, J, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | Deposit date: | 2010-07-13 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
3O6Y
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | Descriptor: | Retro-Aldolase, SULFATE ION | | Authors: | Althoff, E.A, Wang, L, Jiang, L, Moody, J, Bolduc, J, Lassila, J.K, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | Deposit date: | 2010-07-29 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
7TA5
 
 | |
2O5Z
 
 | |
7UQV
 
 | | Pseudobacteroides cellulosolvens pseudo-CphB | | Descriptor: | Cyanophycinase | | Authors: | Sharon, I, Schmeing, T.M. | | Deposit date: | 2022-04-20 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of cyanophycinase in complex with a cyanophycin degradation intermediate.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
7UQW
 
 | | PCC6803 Cyanophycinase S132DAP covalently bound to cyanophycin dimer | | Descriptor: | (2~{S})-4-[[(2~{S})-5-[[azanyl($l^{4}-azanylidene)methyl]amino]-1-$l^{1}-oxidanyl-1-oxidanylidene-pentan-2-yl]amino]-2-$l^{2}-azanyl-4-oxidanylidene-butanoic acid, Cyanophycinase, FORMAMIDE, ... | | Authors: | Sharon, I, Schmeing, T.M. | | Deposit date: | 2022-04-20 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of cyanophycinase in complex with a cyanophycin degradation intermediate.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
2O5X
 
 | |
2O5Y
 
 | |
1C5B
 
 | | DECARBOXYLASE CATALYTIC ANTIBODY 21D8 UNLIGANDED FORM | | Descriptor: | CHIMERIC DECARBOXYLASE ANTIBODY 21D8 | | Authors: | Hotta, K, Wilson, I.A. | | Deposit date: | 1999-11-08 | | Release date: | 2000-10-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalysis of decarboxylation by a preorganized heterogeneous microenvironment: crystal structures of abzyme 21D8.
J.Mol.Biol., 302, 2000
|
|
1C5C
 
 | | DECARBOXYLASE CATALYTIC ANTIBODY 21D8-HAPTEN COMPLEX | | Descriptor: | 2-ACETYLAMINO-NAPTHALENE-1,5-DISULFONIC ACID, CHIMERIC DECARBOXYLASE ANTIBODY 21D8, GLYCEROL | | Authors: | Hotta, K, Wilson, I.A. | | Deposit date: | 1999-11-09 | | Release date: | 2000-10-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Catalysis of decarboxylation by a preorganized heterogeneous microenvironment: crystal structures of abzyme 21D8.
J.Mol.Biol., 302, 2000
|
|
1C1E
 
 | | CRYSTAL STRUCTURE OF A DIELS-ALDERASE CATALYTIC ANTIBODY 1E9 IN COMPLEX WITH ITS HAPTEN | | Descriptor: | 1,7,8,9,10,10-HEXACHLORO-4-METHYL-4-AZA-TRICYCLO[5.2.1.0(2,6)]DEC-8-ENE-3,5-DIONE, CATALYTIC ANTIBODY 1E9 (HEAVY CHAIN), CATALYTIC ANTIBODY 1E9 (LIGHT CHAIN), ... | | Authors: | Xu, J, Wilson, I.A. | | Deposit date: | 1999-07-22 | | Release date: | 2000-03-01 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of shape complementarity and catalytic efficiency from a primordial antibody template.
Science, 286, 1999
|
|
8FXI
 
 | | Cryo-EM structure of Stanieria sp. CphA2 in complex with ADPCP and 4x(beta-Asp-Arg) | | Descriptor: | 1-[(4-aminopyrimidin-5-yl)amino]-2,5-anhydro-1-deoxy-6-O-[(S)-hydroxy{[(R)-hydroxy(phosphonomethyl)phosphoryl]oxy}phosphoryl]-D-allitol, 4x(beta-Asp-Arg), MAGNESIUM ION, ... | | Authors: | Markus, L.M, Sharon, I, Strauss, M, Schmeing, T.M. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and function of a hexameric cyanophycin synthetase 2.
Protein Sci., 32, 2023
|
|
8FXH
 
 | |
5OJA
 
 | | Structure of MbQ | | Descriptor: | IMIDAZOLE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.347 Å) | | Cite: | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
5OJB
 
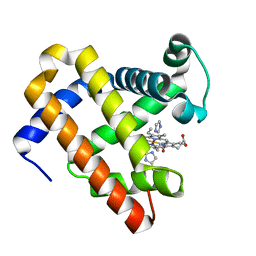 | | Structure of MbQ NMH | | Descriptor: | IMIDAZOLE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
5OJC
 
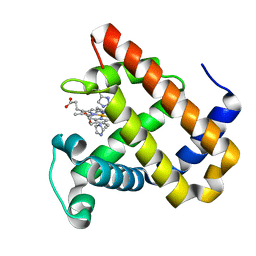 | | Structure of MbQ2.1 NMH | | Descriptor: | IMIDAZOLE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
5OJ9
 
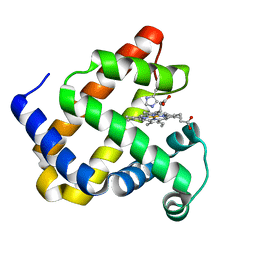 | | Structure of Mb NMH | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
8FE1
 
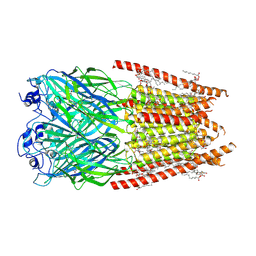 | | Alpha1/BetaB Heteromeric Glycine Receptor in 1 mM Glycine 20 uM Ivermectin State | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gibbs, E, Chakrapani, S. | | Deposit date: | 2022-12-05 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational transitions and allosteric modulation in a heteromeric glycine receptor.
Nat Commun, 14, 2023
|
|
2GTV
 
 | |
2GJZ
 
 | |
