1P5J
 
 | | Crystal Structure Analysis of Human Serine Dehydratase | | Descriptor: | L-serine dehydratase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sun, L, Liu, Y, Rao, Z. | | Deposit date: | 2003-04-27 | | Release date: | 2004-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallization and preliminary crystallographic analysis of human serine dehydratase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1P72
 
 | | Crystal structure of EHV4-TK complexed with Thy and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, THYMIDINE, ... | | Authors: | Gardberg, A, Shuvalova, L, Monnerjahn, C, Konrad, M, Lavie, A. | | Deposit date: | 2003-04-30 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the dual thymidine and thymidylate kinase activity of herpes thymidine kinases.
Structure, 11, 2003
|
|
6G6X
 
 | | 14-3-3sigma in complex with a P129beta3P mutated YAP pS127 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, SODIUM ION, ... | | Authors: | Andrei, S.A, Thijssen, V, Brunsveld, L, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-04-03 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | A study on the effect of synthetic alpha-to-beta3-amino acid mutations on the binding of phosphopeptides to 14-3-3 proteins.
Chem.Commun.(Camb.), 55, 2019
|
|
1P6V
 
 | | Crystal structure of the tRNA domain of transfer-messenger RNA in complex with SmpB | | Descriptor: | 45-MER, SsrA-binding protein | | Authors: | Gutmann, S, Haebel, P.W, Metzinger, L, Sutter, M, Felden, B, Ban, N. | | Deposit date: | 2003-04-30 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB
Nature, 424, 2003
|
|
1P7J
 
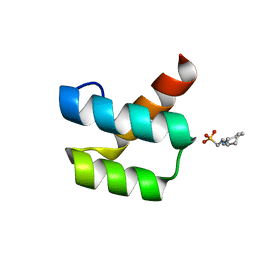 | | Crystal structure of engrailed homeodomain mutant K52E | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Segmentation polarity homeobox protein engrailed | | Authors: | Stollar, E.J, Mayor, U, Lovell, S.C, Federici, L, Freund, S.M, Fersht, A.R, Luisi, B.F. | | Deposit date: | 2003-05-02 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Engrailed Homeodomain Mutants: IMPLICATIONS FOR STABILITY AND DYNAMICS
J.Biol.Chem., 278, 2003
|
|
8SV9
 
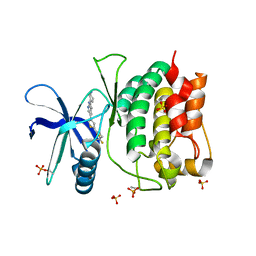 | | Crystal structure of ULK1 kinase domain with inhibitor MR-2088 | | Descriptor: | (4P)-4-[(2P)-2-(1,2,5,6-tetrahydropyridin-3-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]-N-(2,2,2-trifluoroethyl)thiophene-2-carboxamide, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Schonbrunn, E, Sun, L. | | Deposit date: | 2023-05-15 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of potent and selective ULK1/2 inhibitors based on 7-azaindole scaffold with favorable in vivo properties.
Eur.J.Med.Chem., 266, 2024
|
|
6G8A
 
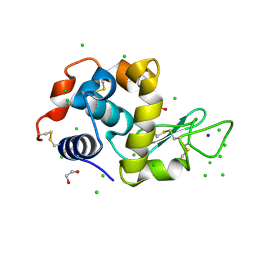 | | Lysozyme solved by Native SAD from a dataset collected in 5 seconds at 1 A wavelength with JUNGFRAU detector | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Leonarski, F, Olieric, V, Vera, L, Redford, S, Wang, M. | | Deposit date: | 2018-04-08 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.143 Å) | | Cite: | Fast and accurate data collection for macromolecular crystallography using the JUNGFRAU detector.
Nat. Methods, 15, 2018
|
|
6G8J
 
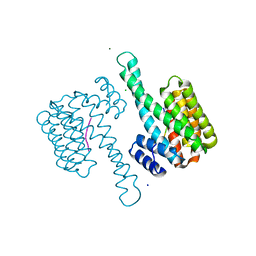 | | 14-3-3sigma in complex with a A130beta3A mutated YAP pS127 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, ACE-ARG-ALA-HIS-SEP-SER-PRO-BAL-SER-LEU-GLN, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Thijssen, V, Brunsveld, L, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A study on the effect of synthetic alpha-to-beta3-amino acid mutations on the binding of phosphopeptides to 14-3-3 proteins.
Chem.Commun.(Camb.), 55, 2019
|
|
8CAR
 
 | | Discovery of the lanthipeptide Curvocidin and structural insights into its trifunctional synthetase CuvL | | Descriptor: | NITRATE ION, PHOSPHATE ION, Serine/threonine protein kinase | | Authors: | Martins, B.M, Sigurdsson, A, Duettmann, A.A, Jasyk, M, Dimos-Roehl, B, Schoepf, F, Gemander, M, Knittel, C.H, Schegotzki, R, Schmid, B, Kosol, S, Pommerening, L, Gonzalez-Viegas, M, Seidel, M, Huegelland, M, Leimkuehler, S, Dobbek, H, Mainz, A, Suessmuth, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery of the Lanthipeptide Curvocidin and Structural Insights into its Trifunctional Synthetase CuvL.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8W6Z
 
 | | Substrate-bound crystal structure of a P450 enzyme DmlH that catalyze intramolecular phenol coupling in the biosynthesis of cihanmycins | | Descriptor: | (E)-3-[2-[(2R,3S)-3-[(1R)-1-aminocarbonyloxypropyl]oxiran-2-yl]phenyl]prop-2-enoic acid, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fang, C, Zhang, L, Zhu, Y, Zhang, C. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate-bound crystal structure of a P450 enzyme DmlH that catalyze intramolecular phenol coupling in the biosynthesis of cihanmycins
To Be Published
|
|
6G8I
 
 | | 14-3-3sigma in complex with a R124beta3R mutated YAP pS127 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, ALA-HIS-SEP-SER-PRO-ALA-SER-LEU-GLN, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Thijssen, V, Brunsveld, L, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A study on the effect of synthetic alpha-to-beta3-amino acid mutations on the binding of phosphopeptides to 14-3-3 proteins.
Chem.Commun.(Camb.), 55, 2019
|
|
3V0F
 
 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 241-576(C363S), form II | | Descriptor: | PHOSPHATE ION, Voltage-sensor containing phosphatase | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6G8Q
 
 | | 14-3-3sigma in complex with a A130beta3A and Q133beta3Q mutated YAP pS127 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Thijssen, V, Brunsveld, L, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A study on the effect of synthetic alpha-to-beta3-amino acid mutations on the binding of phosphopeptides to 14-3-3 proteins.
Chem.Commun.(Camb.), 55, 2019
|
|
3V0T
 
 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Perakine Reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.333 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
1D5A
 
 | | CRYSTAL STRUCTURE OF AN ARCHAEBACTERIAL DNA POLYMERASE D.TOK. DEPOSITION OF SECOND NATIVE STRUCTURE AT 2.4 ANGSTROM | | Descriptor: | MAGNESIUM ION, PROTEIN (DNA POLYMERASE), SULFATE ION | | Authors: | Zhao, Y, Jeruzalmi, D, Leighton, L, Lasken, R, Kuriyan, J. | | Deposit date: | 1999-10-06 | | Release date: | 2000-03-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an archaebacterial DNA polymerase.
Structure Fold.Des., 7, 1999
|
|
1P8D
 
 | | X-Ray Crystal Structure of LXR Ligand Binding Domain with 24(S),25-epoxycholesterol | | Descriptor: | 17-[3-(3,3-DIMETHYL-OXIRANYL)-1-METHYL-PROPYL]-10,13-DIMETHYL-2,3,4,7,8,9,10,11,12,13,14,15,16,17-TETRADECAHYDRO-1H-CYC LOPENTA[A]PHENANTHREN-3-OL, Oxysterols receptor LXR-beta, nuclear receptor coactivator 1 isoform 3 | | Authors: | Williams, S, Bledsoe, R.K, Collins, J.L, Boggs, S, Lambert, M.H, Miller, A.B, Moore, J, McKee, D.D, Moore, L, Nichols, J, Parks, D, Watson, M, Wisely, B, Willson, T.M. | | Deposit date: | 2003-05-06 | | Release date: | 2003-07-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystal structure of the liver X receptor beta ligand binding domain: regulation by
a histidine-tryptophan switch.
J.Biol.Chem., 278, 2003
|
|
8SKD
 
 | |
8CLJ
 
 | | TFIIIC TauB-DNA dimer | | Descriptor: | General transcription factor 3C polypeptide 1, General transcription factor 3C polypeptide 2, General transcription factor 3C polypeptide 4, ... | | Authors: | Seifert-Davila, W, Girbig, M, Hauptmann, L, Hoffmann, T, Eustermann, S, Mueller, C.W. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into human TFIIIC promoter recognition.
Sci Adv, 9, 2023
|
|
8SKX
 
 | |
3UR4
 
 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Dong, A, Dombrovski, L, Senisterra, G, Wernimont, A, Wasney, G.A, Allali Hassani, A, Nguyen, K.T, Smil, D, Bolshan, Y, Hajian, T, Poda, G, Chau, I, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Brown, P, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-21 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
1P93
 
 | | CRYSTAL STRUCTURE OF THE AGONIST FORM OF GLUCOCORTICOID RECEPTOR | | Descriptor: | DEXAMETHASONE, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | Kauppi, B, Jakob, C, Farnegardh, M, Yang, J, Ahola, H, Alarcon, M, Calles, K, Engstrom, O, Harlan, J, Muchmore, S, Ramqvist, A.-K, Thorell, S, Ohman, L, Greer, J, Gustafsson, J.-A, Carlstedt-Duke, J, Carlquist, M. | | Deposit date: | 2003-05-09 | | Release date: | 2003-07-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Three-dimensional Structures of Antagonistic and Agonistic Forms of the Glucocorticoid Receptor Ligand-binding Domain:
RU-486 INDUCES A TRANSCONFORMATION THAT LEADS TO ACTIVE ANTAGONISM.
J.Biol.Chem., 278, 2003
|
|
4BD1
 
 | | Neutron structure of a perdeuterated Toho-1 R274N R276N double mutant Beta-lactamase in complex with a fully deuterated boronic acid (BZB) | | Descriptor: | BENZO[B]THIOPHENE-2-BORONIC ACID, TOHO-1 BETA-LACTAMASE | | Authors: | Tomanicek, S.J, Weiss, K.L, Standaert, R.F, Ostermann, A, Schrader, T.E, Ng, J.D, Coates, L. | | Deposit date: | 2012-10-04 | | Release date: | 2013-01-16 | | Last modified: | 2017-03-22 | | Method: | NEUTRON DIFFRACTION (2.002 Å) | | Cite: | Neutron and X-Ray Crystal Structures of a Perdeuterated Enzyme Inhibitor Complex Reveal the Catalytic Proton Network of the Toho-1 Beta-Lactamase for the Acylation Reaction.
J.Biol.Chem., 288, 2013
|
|
6GDT
 
 | |
5NII
 
 | | Crystal structure of the atypical thioredoxin reductase TRi from Desulfovibrio vulgaris Hildenborough | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Valette, O, Tran, T.T.I, Cavazza, C, Caudeville, E, Brasseur, G, Dolla, A, Talla, E, Pieulle, L. | | Deposit date: | 2017-03-24 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical Function, Molecular Structure and Evolution of an Atypical Thioredoxin Reductase from Desulfovibrio vulgaris.
Front Microbiol, 8, 2017
|
|
6GA2
 
 | | Bacteriorhodopsin, dark state, cell 2 | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
