8HEH
 
 | | Crystal structure of GCN5-related N-acetyltransferase 05790 | | Descriptor: | COENZYME A, GLYCEROL, GNAT family N-acetyltransferase | | Authors: | Xu, M.X, Ran, T.T, Wang, W. | | Deposit date: | 2022-11-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of prodigiosin binding protein PgbP, a GNAT family protein, in Serratia marcescens FS14.
Biochem.Biophys.Res.Commun., 640, 2022
|
|
8YZD
 
 | | Structure of JN.1 RBD protein in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,fusion protein | | Authors: | Wang, Y.J, Zhang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
8YZE
 
 | | The JN.1 spike protein (S) in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,Expression Tag | | Authors: | Wang, Y.J, Zhang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
8YZC
 
 | | Structure of BA.2.86 spike protein in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,Expression Tag | | Authors: | Wang, Y.J, Zang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
8YZB
 
 | | BA.2.86 RBD protein in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,fusion protein | | Authors: | Wang, Y.J, Zhang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
4DRA
 
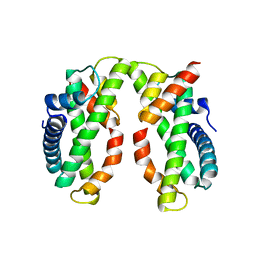 | | Crystal structure of MHF complex | | Descriptor: | Centromere protein S, Centromere protein X | | Authors: | Tao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.414 Å) | | Cite: | The structure of the FANCM-MHF complex reveals physical features for functional assembly
Nat Commun, 3, 2012
|
|
8JEI
 
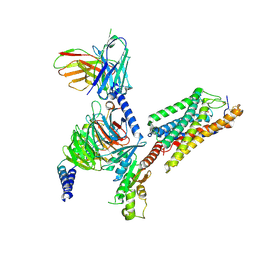 | | Cryo-EM Structure of the compuond 5c-HCAR3-Gi complex | | Descriptor: | 3-nitro-4-(propylamino)benzoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Fang, Y, Pan, X. | | Deposit date: | 2023-05-15 | | Release date: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis for ligand recognition of the human hydroxycarboxylic acid receptor HCAR3.
Cell Rep, 43, 2024
|
|
8JEF
 
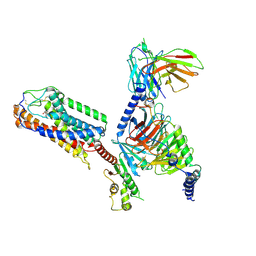 | | Cryo-EM Structure of the 3HO-HCAR3-Gi complex | | Descriptor: | (3S)-3-hydroxyoctanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Fang, Y, Pan, X. | | Deposit date: | 2023-05-15 | | Release date: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis for ligand recognition of the human hydroxycarboxylic acid receptor HCAR3.
Cell Rep, 43, 2024
|
|
5IZQ
 
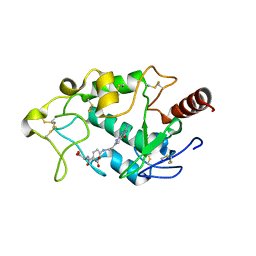 | | Crystal structure of human folate receptor alpha in complex with novel antifolate AGF183 | | Descriptor: | Folate receptor alpha, N-(4-{[2-(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-6-yl)ethyl]amino}benzene-1-carbonyl)-L-glutamic acid | | Authors: | Ke, J, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-03-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Tumor Targeting with Novel 6-Substituted Pyrrolo [2,3-d] Pyrimidine Antifolates with Heteroatom Bridge Substitutions via Cellular Uptake by Folate Receptor alpha and the Proton-Coupled Folate Transporter and Inhibition of de Novo Purine Nucleotide Biosynthesis.
J.Med.Chem., 59, 2016
|
|
7E6T
 
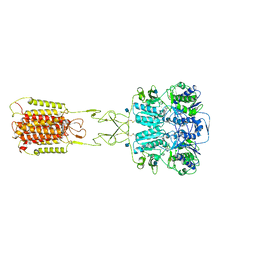 | | Structural insights into the activation of human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYCLOMETHYLTRYPTOPHAN, ... | | Authors: | Geng, Y, Chen, X.C, Wang, L, Cui, Q.Q, Ding, Z.Y, Han, L, Kou, Y.J, Zhang, W.Q, Wang, H.N, Jia, X.M, Dai, M, Shi, Z.Z, Li, Y.Y, Li, X.Y. | | Deposit date: | 2021-02-24 | | Release date: | 2021-09-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the activation of human calcium-sensing receptor.
Elife, 10, 2021
|
|
7E6U
 
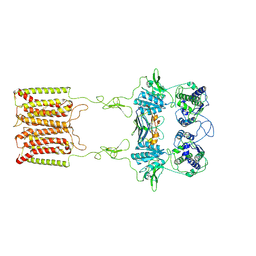 | | the complex of inactive CaSR and NB2D11 | | Descriptor: | Extracellular calcium-sensing receptor, NB-2D11 | | Authors: | Geng, Y, Chen, X.C, Wang, L, Cui, Q.Q, Ding, Z.Y, Han, L, Kou, Y.J, Zhang, W.Q, Wang, H.N, Jia, X.M, Dai, M, Shi, Z.Z, Li, Y.Y, Li, X.Y. | | Deposit date: | 2021-02-24 | | Release date: | 2021-09-22 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural insights into the activation of human calcium-sensing receptor.
Elife, 10, 2021
|
|
7XUF
 
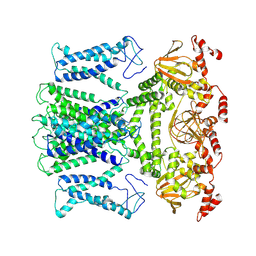 | | Cryo-EM structure of the AKT1-AtKC1 complex from Arabidopsis thaliana | | Descriptor: | POTASSIUM ION, Potassium channel AKT1, Potassium channel KAT3 | | Authors: | Yang, G.H, Lu, Y.M, Jia, Y.T, Yang, F, Zhang, Y.M, Xu, X, Li, X.M, Lei, J.L. | | Deposit date: | 2022-05-18 | | Release date: | 2022-11-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
8ZET
 
 | | Tp-PSI-FCPI-S in Thalassiosira pseudonana | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | Authors: | Feng, Y, Li, Z, Shen, J.R, Wang, W. | | Deposit date: | 2024-05-06 | | Release date: | 2024-12-25 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of PSI-FCPI from Thalassiosira pseudonana grown under high light provide evidence for convergent evolution and light-adaptive strategies in diatom FCPIs.
J Integr Plant Biol, 67, 2025
|
|
7WRH
 
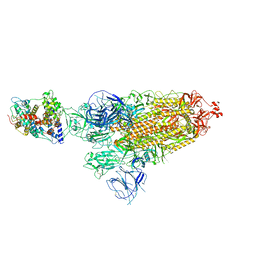 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Han, P, Xie, Y, Qi, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
6B3E
 
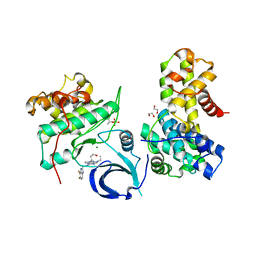 | | Crystal structure of human CDK12/CyclinK in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2S)-1-(6-{[(4,5-difluoro-1H-benzimidazol-2-yl)methyl]amino}-9-ethyl-9H-purin-2-yl)piperidin-2-yl]ethan-1-ol, Cyclin-K, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2017-09-21 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure-Based Design of Selective Noncovalent CDK12 Inhibitors.
ChemMedChem, 13, 2018
|
|
4DRB
 
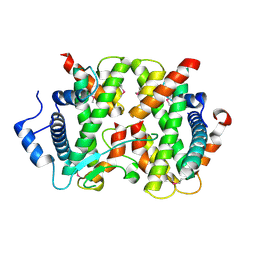 | | The crystal structure of FANCM bound MHF complex | | Descriptor: | Centromere protein S, Centromere protein X, Fanconi anemia group M protein | | Authors: | Tao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | The structure of the FANCM-MHF complex reveals physical features for functional assembly
Nat Commun, 3, 2012
|
|
4MG3
 
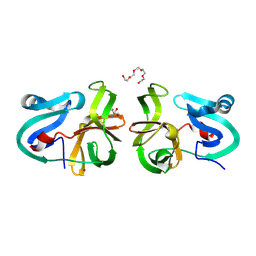 | | Crystal Structural Analysis of 2A Protease from Coxsackievirus A16 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PENTAETHYLENE GLYCOL, Protease 2A, ... | | Authors: | Sun, Y, Wang, X, Dang, M, Yuan, S. | | Deposit date: | 2013-08-28 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | An open conformation determined by a structural switch for 2A protease from coxsackievirus A16.
Protein Cell, 4, 2013
|
|
7WRI
 
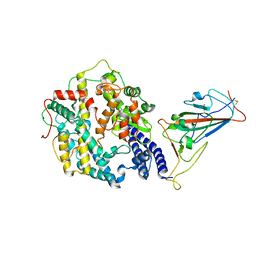 | | Cryo-EM structure of SARS-CoV-2 Omicron spike receptor-binding domain in complex with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Han, P, Xie, Y, Qi, J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-08 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
7WSK
 
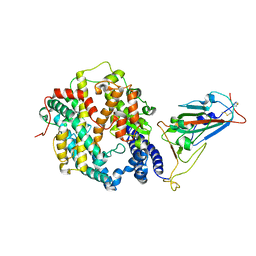 | | Crystal structure of SARS-CoV-2 Omicron spike receptor-binding domain in complex with civet ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Huang, B, Han, P, Qi, J. | | Deposit date: | 2022-01-29 | | Release date: | 2022-06-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
7MXD
 
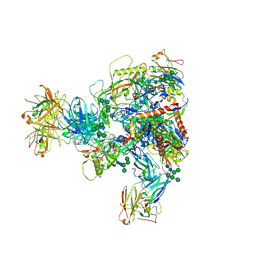 | |
7N28
 
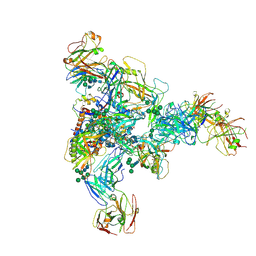 | |
5TSR
 
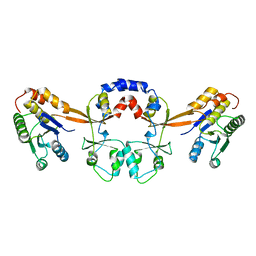 | |
7EWA
 
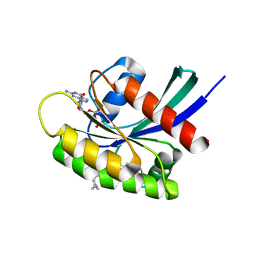 | | GDP-bound KRAS G12D in complex with TH-Z827 | | Descriptor: | 4-[(1~{R},5~{S})-3,8-diazabicyclo[3.2.1]octan-8-yl]-7-(8-methylnaphthalen-1-yl)-2-[[(2~{S})-1-methylpyrrolidin-2-yl]methoxy]-6,8-dihydro-5~{H}-pyrido[3,4-d]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Shen, P, Yang, Y, Yang, Y, Zhang, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | KRAS(G12D) can be targeted by potent inhibitors via formation of salt bridge.
Cell Discov, 8, 2022
|
|
6IHB
 
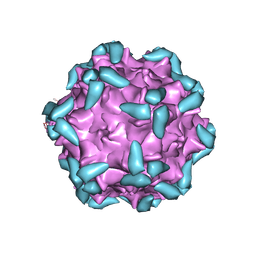 | | Adeno-Associated Virus 2 in complex with AAVR | | Descriptor: | Capsid protein VP1, Dyslexia-associated protein KIAA0319-like protein | | Authors: | Lou, Z.Y, Zhang, R. | | Deposit date: | 2018-09-29 | | Release date: | 2019-03-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Adeno-associated virus 2 bound to its cellular receptor AAVR.
Nat Microbiol, 4, 2019
|
|
7EYK
 
 | |
