7E2Q
 
 | | Crystal structure of Mycoplasma pneumoniae Enolase | | Descriptor: | Enolase, SULFATE ION | | Authors: | Chen, R, Zhang, S, Gan, R, Wang, W, Ran, T, Xiong, Q, Shao, G, Feng, Z. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases.
Front Mol Biosci, 8, 2022
|
|
1T3C
 
 | | Clostridium botulinum type E catalytic domain E212Q mutant | | Descriptor: | CHLORIDE ION, ZINC ION, neurotoxin type E | | Authors: | Agarwal, R, Eswaramoorthy, S, Kumaran, D, Binz, T, Swaminathan, S. | | Deposit date: | 2004-04-26 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of botulinum neurotoxin type E catalytic domain and its mutant Glu212-->Gln reveals the pivotal role of the Glu212 carboxylate in the catalytic pathway
Biochemistry, 43, 2004
|
|
4F3S
 
 | | Crystal structure of periplasmic D-alanine ABC transporter from Salmonella enterica | | Descriptor: | D-ALANINE, GLYCINE, PHOSPHATE ION, ... | | Authors: | Agarwal, R, Chamala, S, Evans, B, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Siedel, R, Villigas, G, Zencheck, W, Foti, R, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-09 | | Release date: | 2012-05-23 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of periplasmic D-alanine ABC transporter from Salmonella enterica
To be Published
|
|
1T61
 
 | | crystal structure of collagen IV NC1 domain from placenta basement membrane | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vanacore, R.M, Shanmugasundararaj, S, Friedman, D.B, Bondar, O, Hudson, B.G, Sundaramoorthy, M. | | Deposit date: | 2004-05-05 | | Release date: | 2004-09-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The alpha1.alpha2 network of collagen IV. Reinforced stabilization of the noncollagenous domain-1 by noncovalent forces and the absence of Met-Lys cross-links
J.Biol.Chem., 279, 2004
|
|
4OHD
 
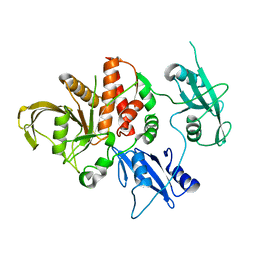 | | LEOPARD Syndrome-Associated SHP2/A461T mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Zhang, R.Y, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L, Liu, S, Zhang, Z.Y. | | Deposit date: | 2014-01-17 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of gain-of-function LEOPARD syndrome-associated SHP2 mutations.
Biochemistry, 53, 2014
|
|
3L47
 
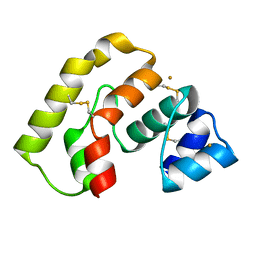 | |
3L4L
 
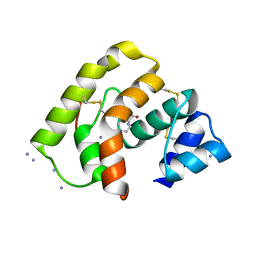 | |
2F1R
 
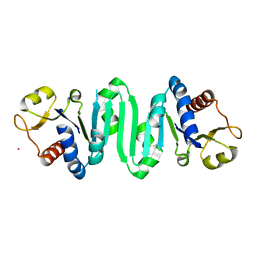 | | Crystal Structure of molybdopterin-guanine biosynthesis protein B (mobB) | | Descriptor: | CHLORIDE ION, PRASEODYMIUM ION, molybdopterin-guanine dinucleotide biosynthesis protein B (mobB) | | Authors: | Damodharan, L, Eswaramoorthy, S, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-11-15 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of molybdopterin-guanine dinucleotide biosynthesis protein B (mobB)
To be Published
|
|
4E3Z
 
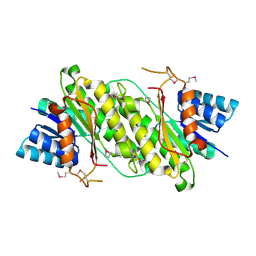 | | Crystal Structure of a oxidoreductase from Rhizobium etli CFN 42 | | Descriptor: | Putative oxidoreductase protein | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-11 | | Release date: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a oxidoreductase from Rhizobium etli CFN 42
To be Published
|
|
1RV9
 
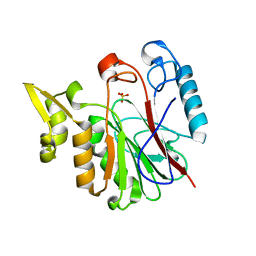 | |
5ZEB
 
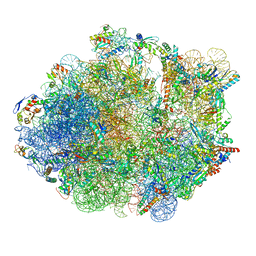 | | M. Smegmatis P/P state 70S ribosome structure | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Mishra, S, Ahmed, T, Tyagi, A, Shi, J, Bhushan, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-09-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA.
Sci Rep, 8, 2018
|
|
5ZEY
 
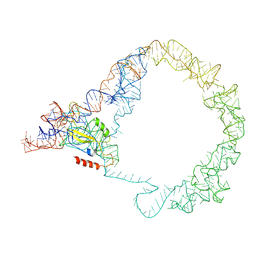 | | M. smegmatis Trans-translation state 70S ribosome | | Descriptor: | A-tRNAfMet, SsrA-binding protein, tmRNA | | Authors: | Mishra, S, Ahmed, T, Tyagi, A, Shi, J, Bhushan, S. | | Deposit date: | 2018-02-28 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA.
Sci Rep, 8, 2018
|
|
1PXR
 
 | | Structure of Pro50Ala mutant of Bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Faham, S, Yang, D, Bare, E, Yohannan, S, Whitelegge, J.P, Bowie, J.U. | | Deposit date: | 2003-07-06 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Side-chain Contributions to Membrane Protein Structure and Stability.
J.Mol.Biol., 335, 2004
|
|
1PY6
 
 | | Bacteriorhodopsin crystallized from bicells | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Faham, S, Yang, D, Bare, E, Yohannan, S, Whitelegge, J.P, Bowie, J.U. | | Deposit date: | 2003-07-08 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Side-chain Contributions to Membrane Protein Structure and Stability.
J.Mol.Biol., 335, 2004
|
|
1RKL
 
 | |
1HSL
 
 | |
3B59
 
 | |
2GVC
 
 | | Crystal structure of flavin-containing monooxygenase (FMO)from S.pombe and substrate (methimazole) complex | | Descriptor: | 1-METHYL-1,3-DIHYDRO-2H-IMIDAZOLE-2-THIONE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Mechanism of action of a flavin-containing monooxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1QAJ
 
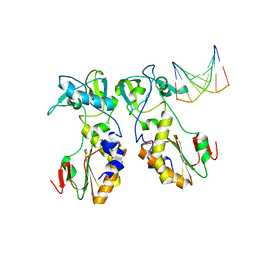 | | CRYSTAL STRUCTURES OF THE N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE COMPLEXED WITH NUCLEIC ACID: FUNCTIONAL IMPLICATIONS FOR TEMPLATE-PRIMER BINDING TO THE FINGERS DOMAIN | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*CP*AP*TP*G)-3'), REVERSE TRANSCRIPTASE | | Authors: | Najmudin, S, Cote, M, Sun, D, Yohannan, S, Montano, S.P, Gu, J, Georgiadis, M.M. | | Deposit date: | 1999-03-18 | | Release date: | 2000-04-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-primer binding to the fingers domain.
J.Mol.Biol., 296, 2000
|
|
3UMA
 
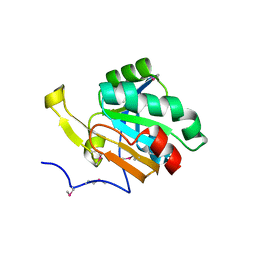 | | Crystal structure of a hypothetical peroxiredoxin protein frm Sinorhizobium meliloti | | Descriptor: | Hypothetical peroxiredoxin protein, SULFATE ION | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-12 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a hypothetical peroxiredoxin protein from Sinorhizobium meliloti
To be Published
|
|
1S0B
 
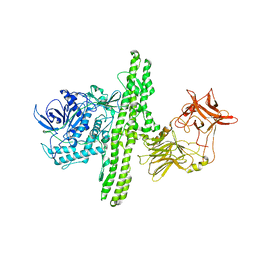 | | Crystal structure of botulinum neurotoxin type B at pH 4.0 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
1S0D
 
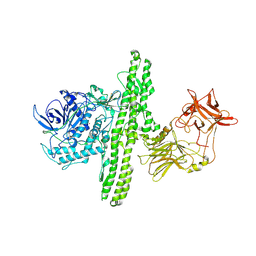 | | Crystal structure of botulinum neurotoxin type B at pH 5.5 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION, ZINC ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
1S0G
 
 | |
2MI1
 
 | |
6GN4
 
 | | tc-DNA/tc-DNA duplex | | Descriptor: | Tc-DNA (5'-D(*(TCJ)P*(TTK)P*(TCJ)P*(TCS)P*(TCS)P*(TCJ)P*(TTK)P*(TTK)P*(TCY)P*(TCJ))-3'), Tc-DNA (5'-D(*(TCS)P*(TTK)P*(TCY)P*(TCY)P*(TCS)P*(TCJ)P*(TCJ)P*(TCS)P*(TCY)P*(TCS))-3') | | Authors: | Istrate, A, Johannsen, S, Istrate, A, Sigel, R.K.O, Leumann, C. | | Deposit date: | 2018-05-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of tricyclo-DNA containing duplexes: insight into enhanced thermal stability and nuclease resistance.
Nucleic Acids Res., 47, 2019
|
|
