5XJO
 
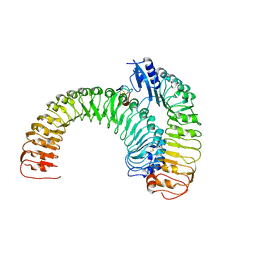 | | Plant receptor ERL1-TMM in complex with peptide EPF1 | | Descriptor: | LRR receptor-like serine/threonine-protein kinase ERL1, Protein EPIDERMAL PATTERNING FACTOR 1, Protein TOO MANY MOUTHS | | Authors: | Chai, J, Lin, G, Zhang, L, Han, Z, Shpak, E.D, Yang, X. | | Deposit date: | 2017-05-03 | | Release date: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (2.626 Å) | | Cite: | A receptor-like protein acts as a specificity switch for the regulation of stomatal development.
Genes Dev., 31, 2017
|
|
1OMC
 
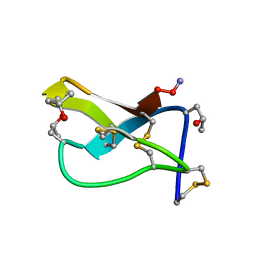 | | SOLUTION STRUCTURE OF OMEGA-CONOTOXIN GVIA USING 2-D NMR SPECTROSCOPY AND RELAXATION MATRIX ANALYSIS | | Descriptor: | OMEGA-CONOTOXIN GVIA | | Authors: | Davis, J.H, Bradley, E.K, Miljanich, G.P, Nadasdi, L, Ramachandran, J, Basus, V.J. | | Deposit date: | 1993-04-28 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of omega-conotoxin GVIA using 2-D NMR spectroscopy and relaxation matrix analysis.
Biochemistry, 32, 1993
|
|
8AZX
 
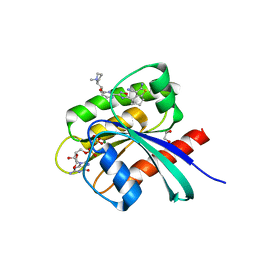 | | KRAS-G12C in complex with BI-2865 | | Descriptor: | (4S)-2-azanyl-4-methyl-4-[3-[4-[(1S)-1-[(2S)-1-methylpyrrolidin-1-ium-2-yl]ethoxy]pyrimidin-2-yl]-1,2,4-oxadiazol-5-yl]-6,7-dihydro-5H-1-benzothiophene-3-carbonitrile, 1,2-ETHANEDIOL, GTPase KRas, ... | | Authors: | Boettcher, J, Herdeis, L. | | Deposit date: | 2022-09-06 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Pan-KRAS inhibitor disables oncogenic signalling and tumour growth.
Nature, 619, 2023
|
|
4B6S
 
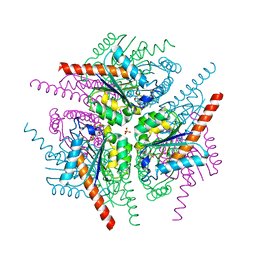 | | Structure of Helicobacter pylori Type II Dehydroquinase inhibited by (2S)-2-Perfluorobenzyl-3-dehydroquinic acid | | Descriptor: | (1R,2S,4S,5R)-2-(2,3,4,5,6-pentafluorophenyl)methyl-1,4,5-trihydroxy-3-oxocyclohexane-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, PHOSPHATE ION | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Lence, E, Tizon, L, Peon, A, Prazeres, V.F.V, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2012-08-14 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic basis of the inhibition of type II dehydroquinase by (2S)- and (2R)-2-benzyl-3-dehydroquinic acids.
ACS Chem. Biol., 8, 2013
|
|
8B00
 
 | | KRAS-G13D in complex with BI-2865 | | Descriptor: | (4S)-2-azanyl-4-methyl-4-[3-[4-[(1S)-1-[(2S)-1-methylpyrrolidin-1-ium-2-yl]ethoxy]pyrimidin-2-yl]-1,2,4-oxadiazol-5-yl]-6,7-dihydro-5H-1-benzothiophene-3-carbonitrile, 1,2-ETHANEDIOL, GTPase KRas, ... | | Authors: | Boettcher, J, Herdeis, L. | | Deposit date: | 2022-09-06 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Pan-KRAS inhibitor disables oncogenic signalling and tumour growth.
Nature, 619, 2023
|
|
1OP9
 
 | | Complex of human lysozyme with camelid VHH HL6 antibody fragment | | Descriptor: | HL6 camel VHH fragment, Lysozyme C | | Authors: | Dumoulin, M, Last, A.M, Desmyter, A, Decanniere, K, Canet, D, Larsson, G, Spencer, A, Archer, D.B, Sasse, J, Muyldermans, S, Wyns, L, Redfield, C, Matagne, A, Robinson, C.V, Dobson, C.M. | | Deposit date: | 2003-03-05 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A camelid antibody fragment inhibits the formation of amyloid fibrils by human lysozyme
Nature, 424, 2003
|
|
4B8U
 
 | | Crystal Structure of 3-hydroxydecanoyl-Acyl Carrier Protein Dehydratase (FabA) from Pseudomonas aeruginosa in complex with N- isobutyl-2-(5-(2-thienyl)-1,2-oxazol-3-yl-)methoxy)acetamide | | Descriptor: | 3-HYDROXYDECANOYL-[ACYL-CARRIER-PROTEIN] DEHYDRATASE, N-isobutyl-2-{[5-(thiophen-2-yl)-1,2-oxazol-3-yl]methoxy}acetamide, SULFATE ION | | Authors: | Moynie, L, McMahon, S.A, Duthie, F.G, Brenk, R, Naismith, J.H. | | Deposit date: | 2012-08-30 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural Insights Into the Mechanism and Inhibition of the Beta-Hydroxydecanoyl-Acyl Carrier Protein Dehydratase from Pseudomonasaeruginosa.
J.Mol.Biol., 425, 2013
|
|
1CY5
 
 | | CRYSTAL STRUCTURE OF THE APAF-1 CARD | | Descriptor: | BETA-MERCAPTOETHANOL, PROTEIN (APOPTOTIC PROTEASE ACTIVATING FACTOR 1), ZINC ION | | Authors: | Vaughn, D.E, Joshua-Tor, L. | | Deposit date: | 1999-08-31 | | Release date: | 1999-09-13 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of Apaf-1 caspase recruitment domain: an alpha-helical Greek key fold for apoptotic signaling.
J.Mol.Biol., 293, 1999
|
|
1P6X
 
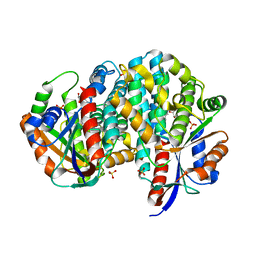 | | Crystal structure of EHV4-TK complexed with Thy and SO4 | | Descriptor: | SULFATE ION, THYMIDINE, Thymidine kinase | | Authors: | Gardberg, A, Shuvalova, L, Monnerjahn, C, Konrad, M, Lavie, A. | | Deposit date: | 2003-04-30 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the dual thymidine and thymidylate kinase activity of herpes thymidine kinases.
Structure, 11, 2003
|
|
8B0K
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli (Apo form) | | Descriptor: | Apolipoprotein N-acyltransferase | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
6ESS
 
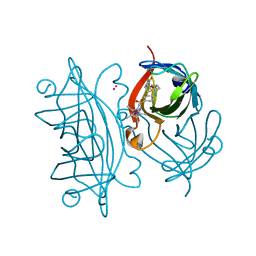 | | Artificial imine reductase mutant S112A-N118P-K121A-S122M | | Descriptor: | IRIDIUM ION, Streptavidin, {N-(4-{[2-(amino-kappaN)ethyl]sulfamoyl-kappaN}phenyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}(chloro)[(1,2,3,4,5-eta)-1,2,3,4,5-pentamethylcyclopentadienyl]iridium(III) | | Authors: | Hestericova, M, Heinisch, T, Alonso-Cotchico, L, Marechal, J.-D, Vidossich, P, Ward, T.R. | | Deposit date: | 2017-10-24 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Directed Evolution of an Artificial Imine Reductase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
4B0I
 
 | | Crystal Structure of 3-hydroxydecanoyl-Acyl Carrier Protein Dehydratase (FabA) mutant (H70N) from Pseudomonas aeruginosa in complex with 3-hydroxydecanoyl-N-acetyl cysteamine | | Descriptor: | 3-HYDROXYDECANOYL-[ACYL-CARRIER-PROTEIN] DEHYDRATASE, S-[2-(acetylamino)ethyl] (3R)-3-hydroxydecanethioate | | Authors: | Moynie, L, McMahon, S.A, Leckie, S.M, Duthie, F.G, Koehnke, A, Naismith, J.H. | | Deposit date: | 2012-07-02 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Insights Into the Mechanism and Inhibition of the Beta-Hydroxydecanoyl-Acyl Carrier Protein Dehydratase from Pseudomonas Aeruginosa
J.Mol.Biol., 425, 2013
|
|
5XPK
 
 | |
6F6W
 
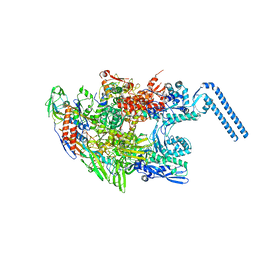 | | Structure of Mycobacterium smegmatis RNA polymerase core | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Kouba, T, Barvik, I, Krasny, L. | | Deposit date: | 2017-12-06 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | The Core and Holoenzyme Forms of RNA Polymerase fromMycobacterium smegmatis.
J. Bacteriol., 201, 2019
|
|
1P8G
 
 | |
6EUN
 
 | | Crystal structure of Neisseria meningitidis vaccine antigen NadA variant 3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ... | | Authors: | Dello Iacono, L, Liguori, A, Malito, E, Bottomley, M.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | NadA3 Structures Reveal Undecad Coiled Coils and LOX1 Binding Regions Competed by Meningococcus B Vaccine-Elicited Human Antibodies.
MBio, 9, 2018
|
|
8AQ2
 
 | | In meso structure of the membrane integral lipoprotein N-acyltransferase Lnt from P. aeruginosa covalently linked with TITC | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, CITRATE ANION, ... | | Authors: | Huang, C.-Y, Weichert, D, Boland, C, Smithers, L, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
4B42
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-07-27 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
8AZZ
 
 | | KRAS-G12V in complex with BI-2865 | | Descriptor: | (4S)-2-azanyl-4-methyl-4-[3-[4-[(1S)-1-[(2S)-1-methylpyrrolidin-1-ium-2-yl]ethoxy]pyrimidin-2-yl]-1,2,4-oxadiazol-5-yl]-6,7-dihydro-5H-1-benzothiophene-3-carbonitrile, 1,2-ETHANEDIOL, GTPase KRas, ... | | Authors: | Boettcher, J, Herdeis, L. | | Deposit date: | 2022-09-06 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Pan-KRAS inhibitor disables oncogenic signalling and tumour growth.
Nature, 619, 2023
|
|
1CW3
 
 | | HUMAN MITOCHONDRIAL ALDEHYDE DEHYDROGENASE COMPLEXED WITH NAD+ AND MN2+ | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, MITOCHONDRIAL ALDEHYDE DEHYDROGENASE, ... | | Authors: | Ni, L, Zhou, J, Hurley, T.D, Weiner, H. | | Deposit date: | 1999-08-25 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Human liver mitochondrial aldehyde dehydrogenase: three-dimensional structure and the restoration of solubility and activity of chimeric forms.
Protein Sci., 8, 1999
|
|
5M6A
 
 | | Crystal structure of cardiotoxic Bence-Jones light chain dimer H9 | | Descriptor: | Bence-Jones light chain, GLYCEROL, PHOSPHATE ION | | Authors: | Oberti, L, Rognoni, P, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-24 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5XAE
 
 | | mutNLIR_LC3B | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-03-12 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between LC3B and RavZ
Biochem. Biophys. Res. Commun., 490, 2017
|
|
3SUX
 
 | | Crystal structure of THF riboswitch, bound with THF | | Descriptor: | 5-HYDROXYMETHYLENE-6-HYDROFOLIC ACID, Riboswitch, SODIUM ION | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Long-range pseudoknot interactions dictate the regulatory response in the tetrahydrofolate riboswitch.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3SUY
 
 | |
5MC8
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-D-glucal and alpha-1,2-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
