2YSF
 
 | | Solution structure of the fourth WW domain from the human E3 ubiquitin-protein ligase Itchy homolog, ITCH | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog | | Authors: | Ohnishi, S, Li, H, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fourth WW domain from the human E3 ubiquitin-protein ligase Itchy homolog, ITCH
To be Published
|
|
6IMA
 
 | | Crystal Structure of ALKBH1 without alpha-1 (N37-C369) | | Descriptor: | CITRIC ACID, Nucleic acid dioxygenase ALKBH1 | | Authors: | Zhang, M, Yang, S, Zhao, W, Li, H. | | Deposit date: | 2018-10-22 | | Release date: | 2020-01-22 | | Last modified: | 2020-04-08 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
6IMC
 
 | | Crystal Structure of ALKBH1 in complex with Mn(II) and N-Oxalylglycine | | Descriptor: | MANGANESE (II) ION, N-OXALYLGLYCINE, Nucleic acid dioxygenase ALKBH1 | | Authors: | Zhang, M, Yang, S, Zhao, W, Li, H. | | Deposit date: | 2018-10-22 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
1S8C
 
 | | Crystal structure of human heme oxygenase in a complex with biliverdine | | Descriptor: | BILIVERDINE IX ALPHA, Heme oxygenase 1 | | Authors: | Lad, L, Friedman, J, Li, H, Bhaskar, B, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2004-02-02 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of Human Heme Oxygenase-1 in a Complex with Biliverdin
Biochemistry, 43, 2004
|
|
1HV8
 
 | |
6U0M
 
 | | Structure of the S. cerevisiae replicative helicase CMG in complex with a forked DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA (15-MER), ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Zhang, D, O'Donnell, M, Li, H. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA unwinding mechanism of a eukaryotic replicative CMG helicase.
Nat Commun, 11, 2020
|
|
6WB9
 
 | | Structure of the S. cerevisiae ER membrane complex | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ... | | Authors: | Bai, L, Li, H. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-03 | | Last modified: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the ER membrane complex, a transmembrane-domain insertase.
Nature, 584, 2020
|
|
6IWF
 
 | | Crystal structure of HitA from Pseudomonas aeruginosa | | Descriptor: | Ferric iron-binding protein HitA | | Authors: | Guo, Y, Zhengrui, Z, Li, H. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.70662332 Å) | | Cite: | Identification and Characterization of a Metalloprotein Involved in Gallium Internalization in Pseudomonas aeruginosa.
Acs Infect Dis., 5, 2019
|
|
7VQX
 
 | | Cryo-EM structure of human vasoactive intestinal polypeptide receptor 2 (VIP2R) in complex with PACAP27 and Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Xu, Y.N, Feng, W.B, Zhou, Q.T, Liang, A.Y, Li, J, Dai, A.T, Zhao, F.H, Yan, J.H, Chen, C.W, Li, H, Zhao, L.H, Xia, T, Jiang, Y, Xu, H.E, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-10-21 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | A distinctive ligand recognition mechanism by the human vasoactive intestinal polypeptide receptor 2.
Nat Commun, 13, 2022
|
|
1SK7
 
 | | Structural Basis for Novel Delta-Regioselective Heme Oxygenation in the Opportunistic Pathogen Pseudomonas aeruginosa | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, hypothetical protein pa-HO | | Authors: | Friedman, J, Lad, L, Li, H, Wilks, A, Poulos, T.L. | | Deposit date: | 2004-03-04 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for novel delta-regioselective heme oxygenation in the opportunistic pathogen Pseudomonas aeruginosa
Biochemistry, 43, 2004
|
|
5YYF
 
 | |
6WGI
 
 | | Atomic model of the mutant OCCM (ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) loaded on DNA at 10.5 A resolution | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (34-MER), ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2YSI
 
 | | Solution structure of the first WW domain from the mouse transcription elongation regulator 1, transcription factor CA150 | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Ohnishi, S, Li, H, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first WW domain from the mouse transcription elongation regulator 1, transcription factor CA150
To be Published
|
|
1JFF
 
 | | Refined structure of alpha-beta tubulin from zinc-induced sheets stabilized with taxol | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lowe, J, Li, H, Downing, K.H, Nogales, E. | | Deposit date: | 2001-06-20 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Refined structure of alpha beta-tubulin at 3.5 A resolution.
J.Mol.Biol., 313, 2001
|
|
1JCI
 
 | | Stabilization of the Engineered Cation-binding Loop in Cytochrome c Peroxidase (CcP) | | Descriptor: | Cytochrome C Peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bhaskar, B, Bonagura, C.A, Li, H, Poulos, T.L. | | Deposit date: | 2001-06-09 | | Release date: | 2002-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cation-induced stabilization of the engineered cation-binding loop in cytochrome c peroxidase (CcP).
Biochemistry, 41, 2002
|
|
5IEU
 
 | | Crystal Structure of Mycobacterium Tuberculosis ATP-independent Proteasome Activator Tetramer | | Descriptor: | Bacterial proteasome activator | | Authors: | Bai, L, Hu, K, Wang, T, Jastrab, J.B, Darwin, K.H, Li, H. | | Deposit date: | 2016-02-25 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of the dodecameric proteasome activator PafE in Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4QYM
 
 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with methionine | | Descriptor: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, MAGNESIUM ION, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-24 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with methionine
To be Published
|
|
5JSJ
 
 | | Crystal structure of Spindlin1 bound to compound EML631 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Spindlin-1, ... | | Authors: | Su, X, Li, H. | | Deposit date: | 2016-05-08 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Developing Spindlin1 small-molecule inhibitors by using protein microarrays
Nat. Chem. Biol., 13, 2017
|
|
5K93
 
 | | PapD wild-type chaperone | | Descriptor: | Chaperone protein PapD | | Authors: | Sarowar, S, Li, H. | | Deposit date: | 2016-05-31 | | Release date: | 2016-07-20 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Escherichia coli P and Type 1 Pilus Assembly Chaperones PapD and FimC Are Monomeric in Solution.
J.Bacteriol., 198, 2016
|
|
4QYR
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsE KS3 | | Descriptor: | ACETIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | Authors: | Kim, Y, Li, H, Endres, M, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4RDP
 
 | | Crystal structure of Cmr4 | | Descriptor: | CRISPR system Cmr subunit Cmr4 | | Authors: | Shao, Y, Tang, L, Li, H. | | Deposit date: | 2014-09-19 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Essential Structural and Functional Roles of the Cmr4 Subunit in RNA Cleavage by the Cmr CRISPR-Cas Complex.
Cell Rep, 9, 2014
|
|
4RDC
 
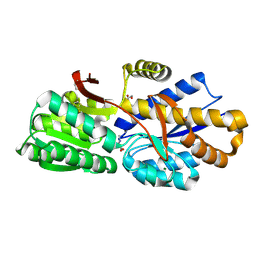 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with proline | | Descriptor: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, FORMIC ACID, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with proline.
To be Published
|
|
4RGP
 
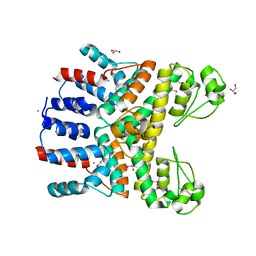 | | Crystal Structure of Uncharacterized CRISPR/Cas System-associated Protein Csm6 from Streptococcus mutans | | Descriptor: | CALCIUM ION, Csm6_III-A, GLYCEROL, ... | | Authors: | Kim, Y, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-24 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Crystal Structure of Uncharacterized CRISPR/Cas System-associated Protein Csm6 from Streptococcus mutans
To be Published
|
|
4RNL
 
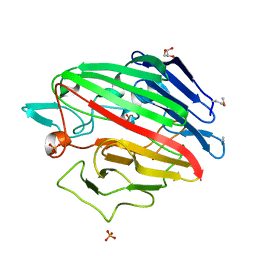 | | The crystal structure of a possible galactose mutarotase from Streptomyces platensis subsp. rosaceus | | Descriptor: | GLYCEROL, PHOSPHATE ION, possible galactose mutarotase | | Authors: | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-26 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a possible galactose mutarotase from Streptomyces platensis subsp. rosaceus
To be Published
|
|
4RR4
 
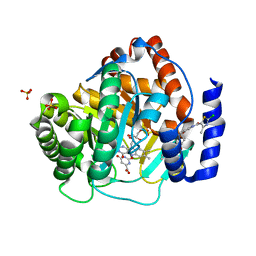 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A367 | | Descriptor: | 2-chloro-N-[3-(4-{[(2Z)-2-cyano-3-cyclopropyl-3-hydroxyprop-2-enoyl]amino}phenoxy)phenyl]-4-methyl-1,3-thiazole-5-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Ren, X, Zhu, J, Li, H. | | Deposit date: | 2014-11-06 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A367
TO BE PUBLISHED
|
|
