4DG1
 
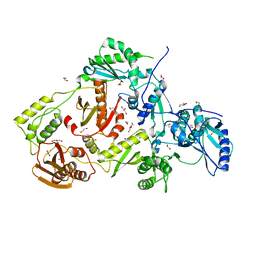 | | Crystal structure of HIV-1 reverse transcriptase (RT) with polymorphism mutation K172A and K173A | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Tu, X, Kirby, K.A, Marchand, B, Sarafianos, S.G. | | Deposit date: | 2012-01-24 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | HIV-1 Reverse Transcriptase (RT) Polymorphism 172K Suppresses the Effect of Clinically Relevant Drug Resistance Mutations to Both Nucleoside and Non-nucleoside RT Inhibitors.
J.Biol.Chem., 287, 2012
|
|
8GTP
 
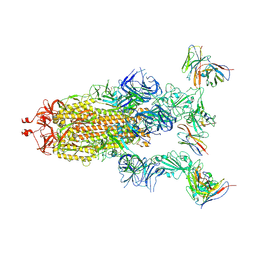 | | cryo-EM structure of Omicron BA.5 S protein in complex with XGv289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, X.Y, Zhang, Y.Y, Chi, X.M, Huang, B.D, Wu, L.S, Zhou, Q. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants.
Cell Discov, 9, 2023
|
|
8GTO
 
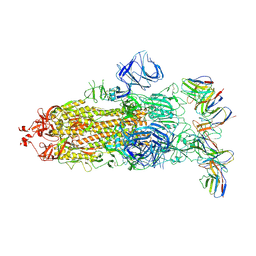 | | cryo-EM structure of Omicron BA.5 S protein in complex with XGv282 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, X.Y, Zhang, Y.Y, Chi, X.M, Huang, B.D, Wu, L.S, Zhou, Q. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants.
Cell Discov, 9, 2023
|
|
8GTQ
 
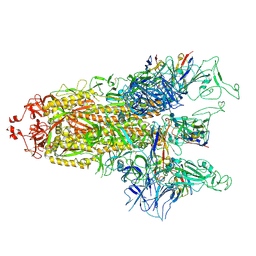 | | cryo-EM structure of Omicron BA.5 S protein in complex with S2L20 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xia, X.Y, Zhang, Y.Y, Chi, X.M, Huang, B.D, Wu, L.S, Zhou, Q. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants.
Cell Discov, 9, 2023
|
|
8HF5
 
 | |
8HF6
 
 | | Cryo-EM structure of nucleotide-bound ComA E647Q mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Competence factor transporting ATP-binding protein/permease ComA | | Authors: | Yu, L, Xin, X, Min, L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of peptide secretion for Quorum sensing by ComA.
Nat Commun, 14, 2023
|
|
8HF4
 
 | |
8K4H
 
 | | Crystal structure of PDE4D complexed with benzbromarone | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, ZINC ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-07-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8K4C
 
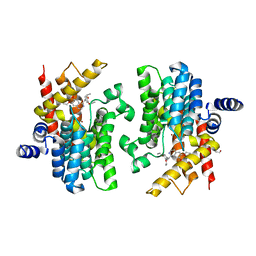 | | Crystal structure of PDE4D complexed with ethaverine hydrochloride | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3,4-diethoxyphenyl)methyl]-6,7-diethoxy-isoquinoline, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8HF7
 
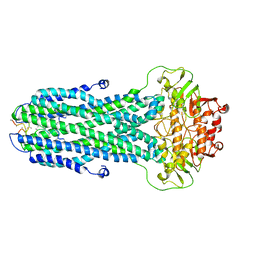 | |
8TL7
 
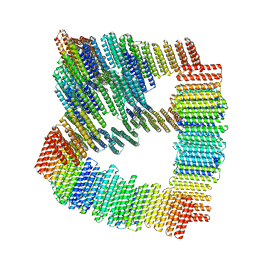 | |
2FVU
 
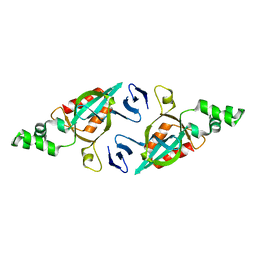 | | Structure of the yeast Sir3 BAH domain | | Descriptor: | Regulatory protein SIR3 | | Authors: | Xu, R.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain.
Mol.Cell.Biol., 26, 2006
|
|
6IMA
 
 | | Crystal Structure of ALKBH1 without alpha-1 (N37-C369) | | Descriptor: | CITRIC ACID, Nucleic acid dioxygenase ALKBH1 | | Authors: | Zhang, M, Yang, S, Zhao, W, Li, H. | | Deposit date: | 2018-10-22 | | Release date: | 2020-01-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
8GQU
 
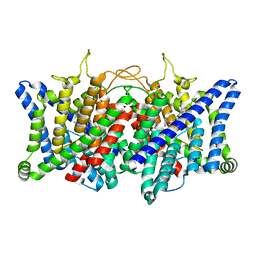 | | AK-42 inhibitor binding human ClC-2 TMD | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-phenylmethoxy-phenyl]amino]pyridine-3-carboxylic acid, Chloride channel protein 2 | | Authors: | Wang, L. | | Deposit date: | 2022-08-30 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42.
Nat Commun, 14, 2023
|
|
4GIG
 
 | |
8HR7
 
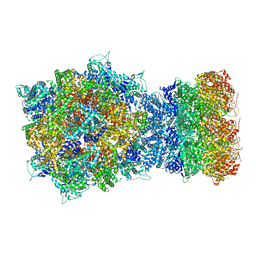 | | Structure of RdrA-RdrB complex | | Descriptor: | Adenosine deaminase, Archaeal ATPase | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HRC
 
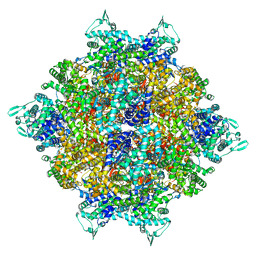 | | Structure of dodecameric RdrB cage | | Descriptor: | Adenosine deaminase | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HR9
 
 | | Structure of tetradecameric RdrA ring | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HR8
 
 | | Structure of heptameric RdrA ring | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HRB
 
 | | Structure of tetradecameric RdrA ring in RNA-loading state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase, RNA (5'-R(*GP*UP*CP*CP*AP*GP*CP*GP*UP*CP*AP*UP*CP*GP*CP*UP*GP*GP*AP*C)-3') | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HRA
 
 | | Structure of heptameric RdrA ring in RNA-loading state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase, RNA (5'-R(P*GP*UP*CP*CP*AP*GP*CP*GP*UP*CP*AP*UP*CP*GP*CP*UP*GP*GP*AP*C)-3') | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8I0C
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S0703 | | Descriptor: | 1-[4-[3,5-bis(chloranyl)phenyl]-3-fluoranyl-phenyl]cyclopropane-1-carboxylic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, He, S, Liu, Y, Fang, P, Sun, H. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
4GH6
 
 | | Crystal structure of the PDE9A catalytic domain in complex with inhibitor 28 | | Descriptor: | High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, N-(4-methoxyphenyl)-N~2~-[1-(2-methylphenyl)-4-oxo-4,5-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl]-L-alaninamide, ... | | Authors: | Hou, J, Ke, H. | | Deposit date: | 2012-08-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Based Discovery of Highly Selective Phosphodiesterase-9A Inhibitors and Implications for Inhibitor Design.
J.Med.Chem., 55, 2012
|
|
6JUE
 
 | | The complex of PDZ and PBM | | Descriptor: | Partitioning defective 3 homolog, SULFATE ION, THR-ILE-ILE-THR-LEU | | Authors: | Liu, Z. | | Deposit date: | 2019-04-13 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Par complex cluster formation mediated by phase separation.
Nat Commun, 11, 2020
|
|
4GY5
 
 | | Crystal structure of the tandem tudor domain and plant homeodomain of UHRF1 with Histone H3K9me3 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Peptide from Histone H3.3, ZINC ION | | Authors: | Cheng, J, Yang, Y, Fang, J, Xiao, J, Zhu, T, Chen, F, Wang, P, Xu, Y. | | Deposit date: | 2012-09-05 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.956 Å) | | Cite: | Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein
J.Biol.Chem., 288, 2013
|
|
