1XNF
 
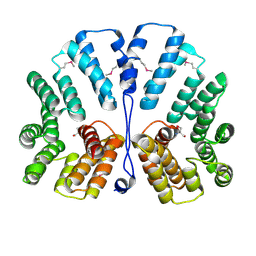 | | Crystal structure of E.coli TPR-protein NlpI | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Lipoprotein nlpI | | Authors: | Wilson, C.G, Kajander, T, Regan, L. | | Deposit date: | 2004-10-04 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The crystal structure of NlpI. A prokaryotic tetratricopeptide repeat protein with a globular fold.
FEBS J., 272, 2005
|
|
1R4K
 
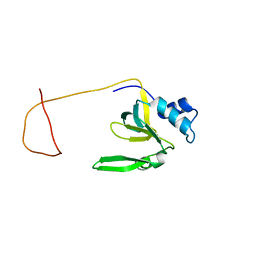 | | Solution Structure of the Drosophila Argonaute 1 PAZ Domain | | Descriptor: | Argonaute 1 | | Authors: | Yan, K.S, Yan, S, Farooq, A, Han, A, Zeng, L, Zhou, M.-M. | | Deposit date: | 2003-10-07 | | Release date: | 2003-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and conserved RNA binding of the PAZ domain
Nature, 426, 2003
|
|
7JXV
 
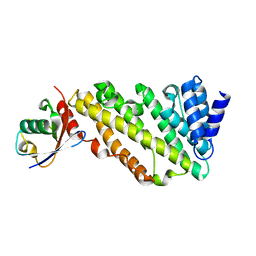 | | ANTH domain of CALM (clathrin-assembly lymphoid myeloid leukemia protein) bound to ubiquitin | | Descriptor: | Phosphatidylinositol-binding clathrin assembly protein, Ubiquitin | | Authors: | Pashkova, N, Gakhar, L, Schnicker, N.J, Piper, R.C. | | Deposit date: | 2020-08-28 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | ANTH domains within CALM, HIP1R, and Sla2 recognize ubiquitin internalization signals.
Elife, 10, 2021
|
|
5DD1
 
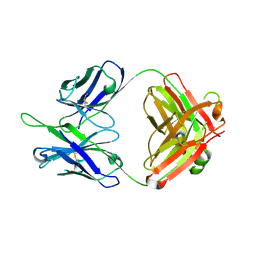 | | Crystal structures in an anti-HIV antibody lineage from immunization of Rhesus macaques | | Descriptor: | ANTI-HIV ANTIBODY DH570 FAB HEAVY CHAIN, ANTI-HIV ANTIBODY DH570 FAB LIGHT CHAIN | | Authors: | Zhang, R, Verkoczy, L, Wiehe, K, Alam, S.M, Nicely, N.I, Santra, S, Bradley, T, Pemble, C, Gao, F, Montefiori, D.C, Bouton-Verville, H, Kelsoe, G, Parks, R, Foulger, A, Tomaras, G, Keple, T.B, Moody, M.A, Liao, H.-X, Haynes, B.F. | | Deposit date: | 2015-08-24 | | Release date: | 2016-05-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Initiation of immune tolerance-controlled HIV gp41 neutralizing B cell lineages.
Sci Transl Med, 8, 2016
|
|
8ONV
 
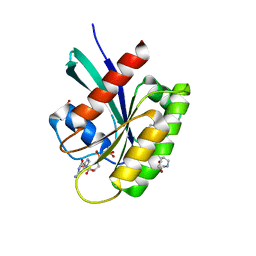 | | KRAS-G13D in complex with BI-2493 | | Descriptor: | (7~{S})-2'-azanyl-3-[2-[(2~{S})-2-methylpiperazin-1-yl]pyrimidin-4-yl]spiro[5,6-dihydro-4~{H}-1,2-benzoxazole-7,4'-6,7-dihydro-5~{H}-1-benzothiophene]-3'-carbonitrile, 1,2-ETHANEDIOL, GTPase KRas, ... | | Authors: | Boettcher, J, Herdeis, L. | | Deposit date: | 2023-04-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Pan-KRAS inhibitor disables oncogenic signalling and tumour growth.
Nature, 619, 2023
|
|
1R4Y
 
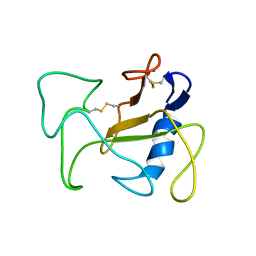 | | SOLUTION STRUCTURE OF THE DELETION MUTANT DELTA(7-22) OF THE CYTOTOXIC RIBONUCLEASE ALPHA-SARCIN | | Descriptor: | Ribonuclease alpha-sarcin | | Authors: | Garcia-Mayoral, M.F, Garcia-Ortega, L, Lillo, M.P, Santoro, J, Martinez Del Pozo, A, Gavilanes, J.G, Rico, M, Bruix, M. | | Deposit date: | 2003-10-09 | | Release date: | 2004-04-06 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the noncytotoxic {alpha}-sarcin mutant {Delta}(7-22): The importance of the native conformation of peripheral loops for activity.
Protein Sci., 13, 2004
|
|
8OEL
 
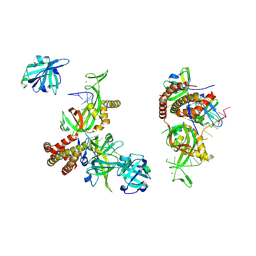 | | Condensed RPA-DNA nucleoprotein filament | | Descriptor: | RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, Replication factor A, ... | | Authors: | Madru, C, Martinez-Carranza, M, Sauguet, L. | | Deposit date: | 2023-03-10 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (8.24 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
6BYY
 
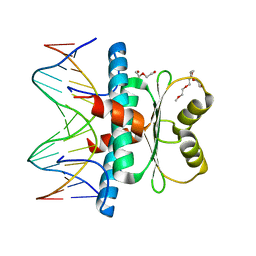 | | MEF2 CHIMERA/DNA Complex | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 3,3',3''-phosphanetriyltripropanoic acid, DNA (5'-D(P*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*GP*A)-3'), ... | | Authors: | Lei, X, Chen, L. | | Deposit date: | 2017-12-21 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Cancer Mutation D83V Induces an alpha-Helix to beta-Strand Conformation Switch in MEF2B.
J. Mol. Biol., 430, 2018
|
|
1RCU
 
 | | X-RAY STRUCTURE OF TM1055 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET VT76 | | Descriptor: | conserved hypothetical protein VT76 | | Authors: | Kuzin, A.P, Chen, Y, Edwards, A, Skarina, T, Savchenko, A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-11-04 | | Release date: | 2003-12-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-RAY STRUCTURE OF TM1055 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET VT76
TO BE PUBLISHED
|
|
6BPG
 
 | |
1XSR
 
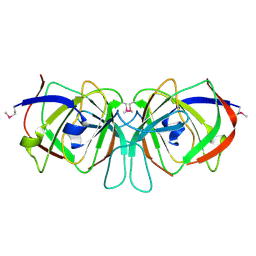 | | X-Ray structure of Northeast Structural Genomics Consortium target SfR7 | | Descriptor: | Ureidoglycolate hydrolase | | Authors: | Kuzin, A.P, Vorobiev, S.M, Edstrom, W, Yong, W, Acton, T, Ma, L.-C, Xiao, R, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-19 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray structure of Northeast
Structural Genomics Consortium target SfR7
To be Published
|
|
5CYG
 
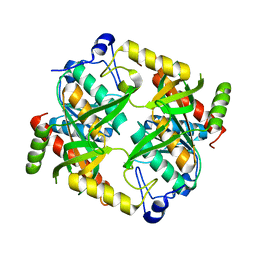 | |
1XN3
 
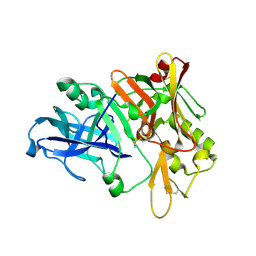 | | Crystal structure of Beta-secretase bound to a long inhibitor with additional upstream residues. | | Descriptor: | Beta-secretase 1, Peptidic inhibitor | | Authors: | Turner III, R.T, Hong, L, Koelsch, G, Ghosh, A.K, Tang, J. | | Deposit date: | 2004-10-04 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural locations and functional roles of new subsites S5, S6, and S7 in memapsin 2 (beta-secretase).
Biochemistry, 44, 2005
|
|
1XUX
 
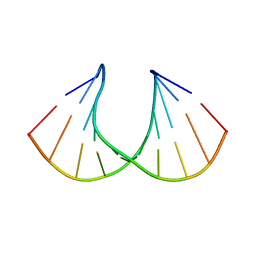 | | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogs | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(NMS)P*AP*CP*GP*C)-3') | | Authors: | Pattanayek, R, Sethaphong, L, Pan, C, Prhavc, M, Prakash, T.P, Manoharan, M, Egli, M. | | Deposit date: | 2004-10-26 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogues.
J.Am.Chem.Soc., 126, 2004
|
|
1RIQ
 
 | | The crystal structure of the catalytic fragment of the alanyl-tRNA synthetase | | Descriptor: | Alanyl-tRNA synthetase | | Authors: | Swairjo, M.A, Otero, F.J, Yang, X.-L, Lovato, M.A, Skene, R.J, McRee, D.E, Ribas de Pouplana, L, Schimmel, P. | | Deposit date: | 2003-11-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Alanyl-tRNA Synthetase Crystal Structure and Design for Acceptor-Stem Recognition
Mol.Cell, 13, 2004
|
|
1XXE
 
 | | RDC refined solution structure of the AaLpxC/TU-514 complex | | Descriptor: | 1,5-ANHYDRO-2-C-(CARBOXYMETHYL-N-HYDROXYAMIDE)-2-DEOXY-3-O-MYRISTOYL-D-GLUCITOL, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Coggins, B.E, McClerren, A.L, Jiang, L, Li, X, Rudolph, J, Hindsgaul, O, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined Solution Structure of the LpxC-TU-514 Complex and pK(a) Analysis of an Active Site Histidine: Insights into the Mechanism and Inhibitor Design
Biochemistry, 44, 2005
|
|
1XS5
 
 | | The Crystal Structure of Lipoprotein Tp32 from Treponema pallidum | | Descriptor: | METHIONINE, Membrane lipoprotein TpN32 | | Authors: | Deka, R.K, Neil, L, Hagman, K.E, Machius, M, Tomchick, D.R, Brautigam, C.A, Norgard, M.V. | | Deposit date: | 2004-10-18 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural evidence that the 32-kilodalton lipoprotein (Tp32) of Treponema pallidum is an L-methionine-binding protein
J.Biol.Chem., 279, 2004
|
|
6C4Q
 
 | | 1.16 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1-100) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Polyketide synthase Pks13 | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-12 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | 1.16 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1-100) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis.
To Be Published
|
|
1XI3
 
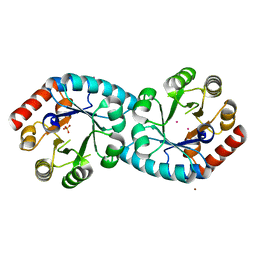 | | Thiamine phosphate pyrophosphorylase from Pyrococcus furiosus Pfu-1255191-001 | | Descriptor: | NICKEL (II) ION, SULFATE ION, Thiamine phosphate pyrophosphorylase, ... | | Authors: | Zhou, W, Chen, L, Tempel, W, Liu, Z.-J, Habel, J, Lee, D, Lin, D, Chang, S.-H, Dillard, B.D, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Kelley, L.-L.C, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thiamine phosphate pyrophosphorylase from Pyrococcus furiosus Pfu-1255191-001
To be published
|
|
6C4Z
 
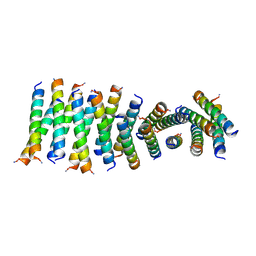 | |
6C55
 
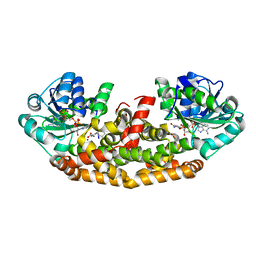 | | Crystal structure of Staphylococcus aureus Ketol-acid Reductosimerrase with hydroxyoxamate inhibitor 3 | | Descriptor: | (cyclohexylamino)(oxo)acetic acid, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, ... | | Authors: | Kandale, A, Patel, K.M, Zheng, S, You, L, Guddat, L.W, Schenk, G, Schmbri, M, McGeary, R.P. | | Deposit date: | 2018-01-14 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductoisomerase (KARI) inhibitors
To Be Published
|
|
8PDN
 
 | | Spiral of assembled human metapneumovirus (HMPV) N-RNA | | Descriptor: | Nucleoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
6C5R
 
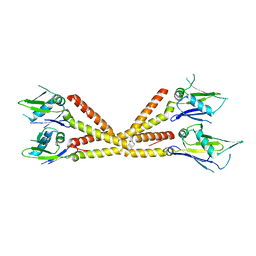 | | Crystal structure of the soluble domain of the mitochondrial calcium uniporter | | Descriptor: | calcium uniporter | | Authors: | Fan, C, Fan, M, Fastman, N, Zhang, J, Feng, L. | | Deposit date: | 2018-01-16 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.09608316 Å) | | Cite: | X-ray and cryo-EM structures of the mitochondrial calcium uniporter.
Nature, 559, 2018
|
|
8PDR
 
 | | Rigid body fit of assembled HMPV N-RNA spiral bound to the C-terminal region of P | | Descriptor: | Nucleoprotein, Phosphoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
8PFH
 
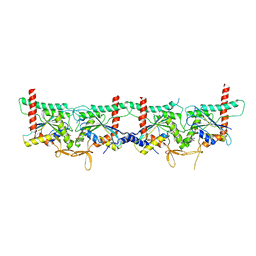 | | Crystal structure of the yeast septin complex Shs1-Cdc12-Cdc3-Cdc10 | | Descriptor: | CDC10 isoform 1, CDC12 isoform 1, Cell division control protein 3, ... | | Authors: | Grupp, B, Denkhaus, L, Gerhardt, S, Gronemeyer, T. | | Deposit date: | 2023-06-16 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | The structure of a tetrameric septin complex reveals a hydrophobic element essential for NC-interface integrity.
Commun Biol, 7, 2024
|
|
