6ITM
 
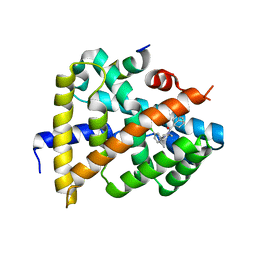 | | Crystal structure of FXR in complex with agonist XJ034 | | Descriptor: | 1-adamantyl-[4-(5-chloranyl-2-methyl-phenyl)piperazin-1-yl]methanone, Bile acid receptor, HD3 Peptide from Nuclear receptor coactivator 1 | | Authors: | Zhang, H, Wang, Z. | | Deposit date: | 2018-11-23 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pose Filter-Based Ensemble Learning Enables Discovery of Orally Active, Nonsteroidal Farnesoid X Receptor Agonists.
J.Chem.Inf.Model., 60, 2020
|
|
7KKZ
 
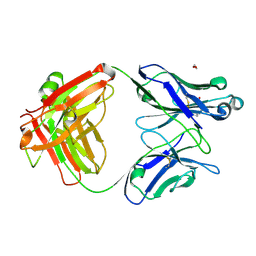 | | Crystal structure of mouse anti-HIV potent neutralizing antibody M4H2K1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Heavy chain of Fab fragment of mouse monoclonal antibody M4H2K1, ... | | Authors: | Kumar, S, Wilson, I.A. | | Deposit date: | 2020-10-28 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Neutralizing Antibodies Induced by First-Generation gp41-Stabilized HIV-1 Envelope Trimers and Nanoparticles.
Mbio, 12, 2021
|
|
7KMD
 
 | |
7KLC
 
 | |
8P0C
 
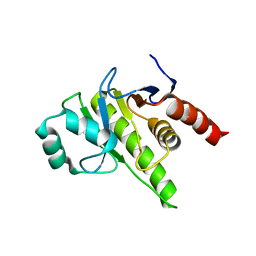 | | Rubella virus p150 macro domain (apo) | | Descriptor: | Non-structural polyprotein p200 | | Authors: | Stoll, G.A, Modis, Y. | | Deposit date: | 2023-05-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure and biochemical activity of the macrodomain from rubella virus p150.
J.Virol., 98, 2024
|
|
8P0E
 
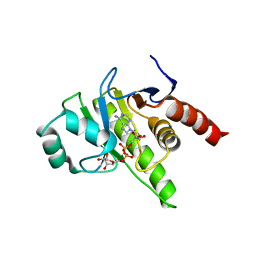 | |
4PSX
 
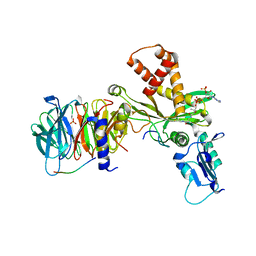 | | Crystal structure of histone acetyltransferase complex | | Descriptor: | COENZYME A, Histone H3, Histone H4, ... | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2014-03-08 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
7E3X
 
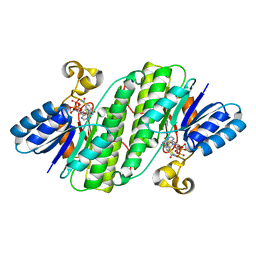 | | Crystal structure of SDR family NAD(P)-dependent oxidoreductase from exiguobacterium | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase | | Authors: | Chen, L, Tang, J, Yuan, S, Zhang, F, Chen, S. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure-guided evolution of a ketoreductase forefficient and stereoselective bioreduction of bulkyalpha-aminobeta-keto esters
Catalysis Science And Technology, 2021
|
|
4PSW
 
 | | Crystal structure of histone acetyltransferase complex | | Descriptor: | COENZYME A, Histone H4 type VIII, Histone acetyltransferase type B catalytic subunit, ... | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2014-03-08 | | Release date: | 2014-07-09 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
3RZ2
 
 | | Crystal of Prl-1 complexed with peptide | | Descriptor: | Prl-1 (PTP4A1), Protein tyrosine phosphatase type IVA 1 | | Authors: | Zhang, Z.-Y, Liu, D, Bai, Y. | | Deposit date: | 2011-05-11 | | Release date: | 2011-10-26 | | Last modified: | 2014-10-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | PRL-1 protein promotes ERK1/2 and RhoA protein activation through a non-canonical interaction with the Src homology 3 domain of p115 Rho GTPase-activating protein.
J.Biol.Chem., 286, 2011
|
|
7KKO
 
 | | Structure of the catalytic domain of tankyrase 1 in complex with olaparib | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase, ZINC ION | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KKM
 
 | | Structure of the catalytic domain of tankyrase 1 | | Descriptor: | Poly [ADP-ribose] polymerase, ZINC ION | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KKN
 
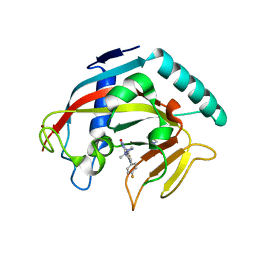 | | Structure of the catalytic domain of tankyrase 1 in complex with talazoparib | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Poly [ADP-ribose] polymerase, ... | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KK3
 
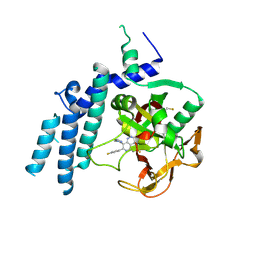 | | Structure of the catalytic domain of PARP1 in complex with talazoparib | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7DU0
 
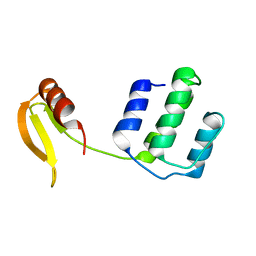 | | Structure of an type I-F anti-crispr protein | | Descriptor: | AcrIF14 | | Authors: | Teng, G, Yue, F. | | Deposit date: | 2021-01-07 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
5VID
 
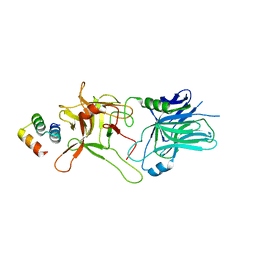 | |
5VLI
 
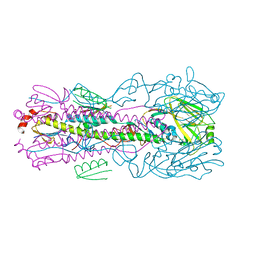 | | Computationally designed inhibitor peptide HB1.6928.2.3 in complex with influenza hemagglutinin (A/PuertoRico/8/1934) | | Descriptor: | 2,5,8,11-TETRAOXATRIDECANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bernard, S.M, Wilson, I.A. | | Deposit date: | 2017-04-25 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Massively parallel de novo protein design for targeted therapeutics.
Nature, 550, 2017
|
|
6JCN
 
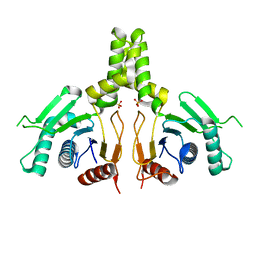 | | Yeast dehydrodolichyl diphosphate synthase complex subunit NUS1 | | Descriptor: | Dehydrodolichyl diphosphate synthase complex subunit NUS1, SULFATE ION | | Authors: | Ko, T.-P, Ma, J, Liu, W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2019-01-29 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural insights to heterodimeric cis-prenyltransferases through yeast dehydrodolichyl diphosphate synthase subunit Nus1.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
1ULW
 
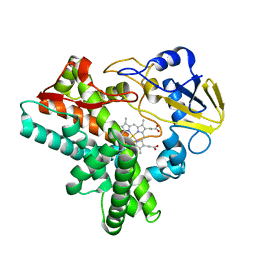 | | Crystal structure of P450nor Ser73Gly/Ser75Gly mutant | | Descriptor: | Cytochrome P450 55A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oshima, R, Fushinobu, S, Su, F, Li, Z, Takaya, N, Shoun, H. | | Deposit date: | 2003-09-16 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for direct hydride transfer from NADH to cytochrome P450nor
J.Mol.Biol., 342, 2004
|
|
7END
 
 | |
7EN9
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-02 | | Descriptor: | 3C-like proteinase, 5-bromanyl-~{N}-methyl-3-nitro-2-[(4~{R},5~{S})-2-(7-oxidanylisoquinolin-4-yl)carbonyl-4-phenyl-2,7-diazaspiro[4.4]nonan-7-yl]benzamide | | Authors: | Hou, N, Peng, C, Hu, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
7EN8
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-04 | | Descriptor: | 3C-like proteinase, GLYCEROL, ~{N}-[(1~{S},2~{R})-2-[[4-bromanyl-2-(methylcarbamoyl)-6-nitro-phenyl]amino]cyclohexyl]isoquinoline-4-carboxamide | | Authors: | Hou, N, Peng, C, Hu, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
7ENE
 
 | |
8Z9C
 
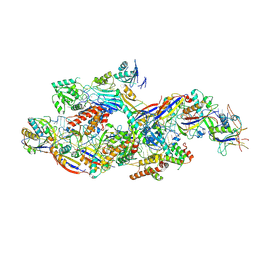 | | Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state I | | Descriptor: | Protein structure, RNA (41-MER), RNA (48-MER), ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z4L
 
 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at substrate-engaged state I | | Descriptor: | RNA (40-MER), RNA (49-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-17 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
