3OO7
 
 | |
6M0Z
 
 | | X-ray structure of Drosophila dopamine transporter with NET-like mutations (D121G/S426M/F471L) in L-norepinephrine bound form | | Descriptor: | Antibody fragment (Fab) 9D5 Light chain, Antibody fragment (Fab) 9D5 heavy chain, CHLORIDE ION, ... | | Authors: | Shabareesh, P, Mallela, A.K, Joseph, D, Penmatsa, A. | | Deposit date: | 2020-02-24 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters.
Nat Commun, 12, 2021
|
|
5D21
 
 | | Multivalency Effects in Glycopeptide Dendrimer Inhibitors of Pseudomonas aeruginosa Biofilms Targeting Lectin LecA | | Descriptor: | CALCIUM ION, LecA, phenyl beta-D-galactopyranoside | | Authors: | Bergmann, M, Michaud, G, Visini, R, Jin, X, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multivalency effects on Pseudomonas aeruginosa biofilm inhibition and dispersal by glycopeptide dendrimers targeting lectin LecA.
Org.Biomol.Chem., 14, 2016
|
|
3OOW
 
 | | Octameric structure of the phosphoribosylaminoimidazole carboxylase catalytic subunit from Francisella tularensis subsp. tularensis SCHU S4. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, F.W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Octameric structure of the phosphoribosylaminoimidazole carboxylase catalytic subunit from Francisella tularensis subsp. tularensis SCHU S4.
To be Published
|
|
3K02
 
 | | Crystal structures of the GacH receptor of Streptomyces glaucescens GLA.O in the unliganded form and in complex with acarbose and an acarbose homolog. Comparison with acarbose-loaded maltose binding protein of Salmonella typhimurium. | | Descriptor: | 4,6-dideoxy-4-{[(1S,2R,3R,4S,5S)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-allopyranosyl-(1->4)-alpha-D-glucopyranosyl-(1->4)-alpha-D-glucopyranosyl-(1->4)-alpha-D-glucopyranose, Acarbose/maltose binding protein GacH, SULFATE ION | | Authors: | Vahedi-Faridi, A, Licht, A, Bulut, H, Schneider, E. | | Deposit date: | 2009-09-24 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structures of the Solute Receptor GacH of Streptomyces glaucescens in Complex with Acarbose and an Acarbose Homolog: Comparison with the Acarbose-Loaded Maltose-Binding Protein of Salmonella typhimurium.
J.Mol.Biol., 397, 2010
|
|
2K4X
 
 | | Solution structure of 30S ribosomal protein S27A from Thermoplasma acidophilum | | Descriptor: | 30S ribosomal protein S27ae, ZINC ION | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Arrowsmith, C, Montelione, G.T, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2008-06-20 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 30S ribosomal protein S27A from Thermoplasma acidophilum/Northeast Structural Genomics Consortium Target TaT88/Ontario Center for Structural Proteomics target ta1093
To be Published
|
|
3JD0
 
 | | Glutamate dehydrogenase in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
4L6X
 
 | |
3JD4
 
 | | Glutamate dehydrogenase in complex with NADH and GTP, closed conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
8G83
 
 | | Structure of NAD+ consuming protein Acinetobacter baumannii TIR domain | | Descriptor: | NAD(+) hydrolase AbTIR | | Authors: | Klontz, E.H, Wang, Y, Glendening, G, Carr, J, Tsibouris, T, Buddula, S, Nallar, S, Soares, A, Snyder, G.A. | | Deposit date: | 2023-02-17 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | The structure of NAD + consuming protein Acinetobacter baumannii TIR domain shows unique kinetics and conformations.
J.Biol.Chem., 299, 2023
|
|
3OP8
 
 | | Crystal structure of the domain V from beta2-glycoprotein I | | Descriptor: | Beta-2-glycoprotein 1, SULFATE ION | | Authors: | Kolyada, A, Lee, C.-J, De Biasio, A, Beglova, N. | | Deposit date: | 2010-08-31 | | Release date: | 2011-01-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Dimeric Inhibitor Targeting Beta2GPI in Beta2GPI/Antibody Complexes Implicated in Antiphospholipid Syndrome.
Plos One, 5, 2010
|
|
3JTI
 
 | | Crystal structure of the complex formed between Phospholipase A2 with beta-amyloid fragment, Lys-Gly-Ala-Ile-Ile-Gly-Leu-Met at 1.8 A resolution | | Descriptor: | CALCIUM ION, Phospholipase A2 isoform 3, octapeptide from Amyloid beta A4 protein | | Authors: | Pandey, N, Mirza, Z, Vikram, G, Singh, N, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-09-12 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex formed between Phospholipase A2 with beta-amyloid fragment, Lys-Gly-Ala-Ile-Ile-Gly-Leu-Met at 1.8 A resolution
To be Published
|
|
1AVH
 
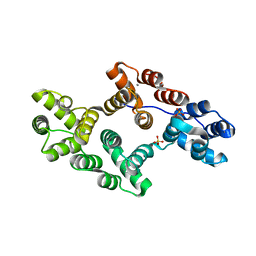 | | CRYSTAL AND MOLECULAR STRUCTURE OF HUMAN ANNEXIN V AFTER REFINEMENT. IMPLICATIONS FOR STRUCTURE, MEMBRANE BINDING AND ION CHANNEL FORMATION OF THE ANNEXIN FAMILY OF PROTEINS | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Huber, R, Berendes, R, Burger, A, Schneider, M, Karshikov, A, Luecke, H, Roemisch, J, Paques, E. | | Deposit date: | 1991-10-17 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal and molecular structure of human annexin V after refinement. Implications for structure, membrane binding and ion channel formation of the annexin family of proteins.
J.Mol.Biol., 223, 1992
|
|
5EU0
 
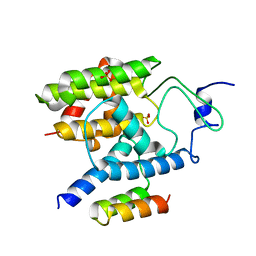 | |
4FIV
 
 | | FIV PROTEASE COMPLEXED WITH AN INHIBITOR LP-130 | | Descriptor: | 4-[2-(2-ACETYLAMINO-3-NAPHTALEN-1-YL-PROPIONYLAMINO)-4-METHYL-PENTANOYLAMINO]-3-HYDROXY-6-METHYL-HEPTANOIC ACID [1-(1-CARBAMOYL-2-NAPHTHALEN-1-YL-ETHYLCARBAMOYL)-PROPYL]-AMIDE, FELINE IMMUNODEFICIENCY VIRUS PROTEASE | | Authors: | Kervinen, J, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-07-15 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a universal inhibitor of retroviral proteases: comparative analysis of the interactions of LP-130 complexed with proteases from HIV-1, FIV, and EIAV.
Protein Sci., 7, 1998
|
|
4L52
 
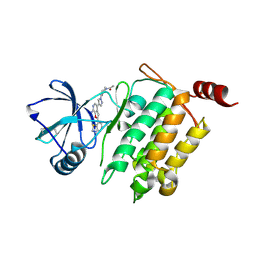 | | Crystal Structure of 1-(4-{4-[7-amino-2-(1,2,3-benzothiadiazol-7-yl)furo[2,3-c]pyridin-4-yl]-1H-pyrazol-1-yl}piperidin-1-yl)ethan-1-one bound to TAK1-TAB1 | | Descriptor: | 1-(4-{4-[7-amino-2-(1,2,3-benzothiadiazol-7-yl)furo[2,3-c]pyridin-4-yl]-1H-pyrazol-1-yl}piperidin-1-yl)ethanone, Mitogen-activated protein kinase kinase kinase 7, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera | | Authors: | Wang, J, Hornberger, K.R, Crew, A.P, Jestel, A, Maskos, K, Moertl, M. | | Deposit date: | 2013-06-10 | | Release date: | 2013-07-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Discovery and optimization of 7-aminofuro[2,3-c]pyridine inhibitors of TAK1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6GOD
 
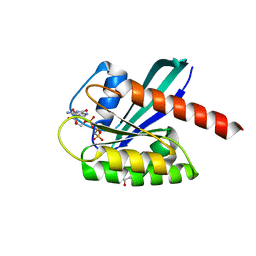 | | KRAS full length wild-type GPPNHP | | Descriptor: | GLYCEROL, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Cruz-Migoni, A, Quevedo, C.E, Carr, S.B, Ehebauer, M.T, Phillips, S.V.E, Rabbitts, T.H. | | Deposit date: | 2018-06-01 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4L6B
 
 | |
3OXP
 
 | | Structure of phosphotransferase enzyme II, A component from Yersinia pestis CO92 at 1.2 A resolution | | Descriptor: | GLYCEROL, Phosphotransferase enzyme II, A component, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-21 | | Release date: | 2010-10-13 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of phosphotransferase enzyme II, A component from Yersinia pestis CO92 at 1.2 A resolution
To be Published
|
|
5EHX
 
 | | Crystal structure of MSF-aged Torpedo californica Acetylcholinesterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, ... | | Authors: | Pesaresi, A, Lamba, D. | | Deposit date: | 2015-10-29 | | Release date: | 2016-11-09 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Disentangling the formation, mechanism, and evolvement of the covalent methanesulfonyl fluoride acetylcholinesterase adduct: Insights into an aged-like inactive complex susceptible to reactivation by a combination of nucleophiles.
Protein Sci., 33, 2024
|
|
3J9M
 
 | | Structure of the human mitochondrial ribosome (class 1) | | Descriptor: | 12S rRNA, 16S rRNA, E-site tRNA, ... | | Authors: | Amunts, A, Brown, A, Toots, J, Scheres, S.H, Ramakrishnan, V. | | Deposit date: | 2015-02-08 | | Release date: | 2015-04-15 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ribosome. The structure of the human mitochondrial ribosome.
Science, 348, 2015
|
|
5EI5
 
 | | Crystal structure of MSF-aged Torpedo californica Acetylcholinesterase in complex with alkylene-linked bis-tacrine dimer (7 carbon linker) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Pesaresi, A, Lamba, D. | | Deposit date: | 2015-10-29 | | Release date: | 2016-11-09 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Disentangling the formation, mechanism, and evolvement of the covalent methanesulfonyl fluoride acetylcholinesterase adduct: Insights into an aged-like inactive complex susceptible to reactivation by a combination of nucleophiles.
Protein Sci., 33, 2024
|
|
3J5Y
 
 | | Structure of the mammalian ribosomal pre-termination complex associated with eRF1-eRF3-GDPNP | | Descriptor: | 5'-R(*AP*UP*UP*GP*UP*AP*AP*AP*AP*A)-3', Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, Eukaryotic peptide chain release factor subunit 1, ... | | Authors: | des Georges, A, Hashem, Y, Unbehaun, A, Grassucci, R.A, Taylor, D, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-25 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Structure of the mammalian ribosomal pre-termination complex associated with eRF1*eRF3*GDPNP.
Nucleic Acids Res., 42, 2014
|
|
4LCO
 
 | | Crystal structure of NE0047 with complex with substrate ammeline | | Descriptor: | 4,6-diamino-1,3,5-triazin-2-ol, Cytidine and deoxycytidylate deaminase zinc-binding region, ZINC ION | | Authors: | Bitra, A, Biswas, A, Anand, R. | | Deposit date: | 2013-06-22 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the substrate specificity of cytidine deaminase superfamily Guanine deaminase
Biochemistry, 52, 2013
|
|
5D4W
 
 | | Crystal structure of Hsp104 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Putative heat shock protein | | Authors: | Heuck, A, Schitter-Sollner, S, Clausen, T. | | Deposit date: | 2015-08-09 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the disaggregase activity and regulation of Hsp104.
Elife, 5, 2016
|
|
