8I3U
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 14B1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 14B1, Light chain of Fab 14B1, ... | | Authors: | Li, Z, Yu, F, Cao, S. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
7D3T
 
 | | Crystal structure of OSPHR2 in complex with DNA | | Descriptor: | DNA (5'-D(P*CP*TP*CP*GP*GP*AP*TP*AP*TP*CP*CP*TP*CP*AP*AP*G)-3'), DNA (5'-D(P*GP*CP*TP*TP*GP*AP*GP*GP*AP*TP*AP*TP*CP*CP*GP*A)-3'), Protein PHOSPHATE STARVATION RESPONSE 2 | | Authors: | Guan, Z.Y, Zhang, Z.F, Liu, Z. | | Deposit date: | 2020-09-20 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2 - PHR2 complex.
Nat Commun, 13, 2022
|
|
8INP
 
 | | A reversible glycosyltransferase of tectorigenin - Bc7OUGT | | Descriptor: | Bc7OUGT, URIDINE-5'-DIPHOSPHATE, beta-D-glucopyranose | | Authors: | Zhang, Z.Y, Lu, L, Guan, Z.F, Cheng, W.J. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Functional characterization and structural basis of a reversible glycosyltransferase involves in plant chemical defence.
Plant Biotechnol J, 21, 2023
|
|
8ITA
 
 | | A reversible glycosyltransferase of tectorigenin - Bc7OUGT complexed with UDP and tectorigenin | | Descriptor: | 3-(4-hydroxyphenyl)-6-methoxy-5,7-bis(oxidanyl)chromen-4-one, Bc7OUGT, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhang, Z.Y, Lu, L, Guan, Z.F, Cheng, W.J. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional characterization and structural basis of a reversible glycosyltransferase involves in plant chemical defence.
Plant Biotechnol J, 21, 2023
|
|
8J6T
 
 | | Cryo-EM structure of the double CAF-1 bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8J6S
 
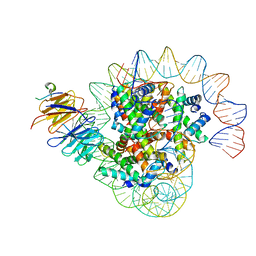 | | Cryo-EM structure of the single CAF-1 bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8IQF
 
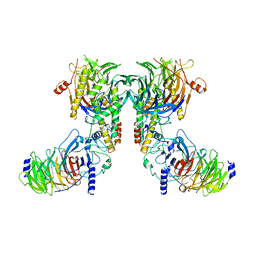 | | Cryo-EM structure of the dimeric human CAF1-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8IQG
 
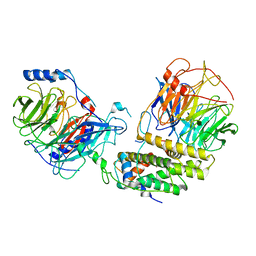 | | Cryo-EM structure of the monomeric human CAF1-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8IDE
 
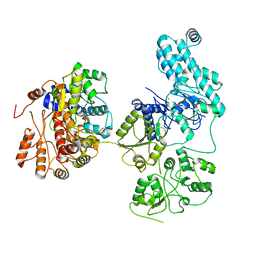 | | Structure of an ancient TsaD-TsaC-SUA5-TcdA modular enzyme (TsaN) | | Descriptor: | MANGANESE (II) ION, N(6)-L-threonylcarbamoyladenine synthase | | Authors: | Zhang, Z.L, Jin, M.Q, Yu, Z.J, Chen, W, Wang, X.L, Lei, D.S, Zhang, W.H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure-function analysis of an ancient TsaD-TsaC-SUA5-TcdA modular enzyme reveals a prototype of tRNA t6A and ct6A synthetases.
Nucleic Acids Res., 51, 2023
|
|
8J63
 
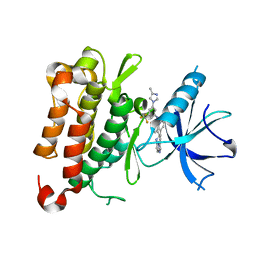 | | The crystal structure of TrkA kinase in complex with 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-11-oxo-5-oxa-10,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Descriptor: | 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-11-oxo-5-oxa-10,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide, High affinity nerve growth factor receptor | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.0005 Å) | | Cite: | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
7C2L
 
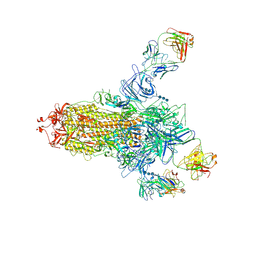 | | S protein of SARS-CoV-2 in complex bound with 4A8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Guo, Y.Y, Li, Y.N, Xia, L, Zhou, Q. | | Deposit date: | 2020-05-08 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2.
Science, 369, 2020
|
|
8JGD
 
 | | GDP-bound KRAS G12C in complex with YK-8S | | Descriptor: | (2~{S})-1-[4-[7-(8-ethynyl-7-fluoranyl-naphthalen-1-yl)-8-fluoranyl-2-[[(2~{R},8~{S})-2-fluoranyl-1,2,3,5,6,7-hexahydropyrrolizin-8-yl]methoxy]pyrido[4,3-d]pyrimidin-4-yl]piperazin-1-yl]-2-oxidanyl-propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Z.M, Wang, R.L. | | Deposit date: | 2023-05-20 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.60037053 Å) | | Cite: | Simultaneous Covalent Modification of K-Ras(G12D) and K-Ras(G12C) with Tunable Oxirane Electrophiles.
J.Am.Chem.Soc., 145, 2023
|
|
8JHL
 
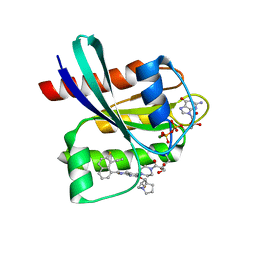 | | GDP-bound KRAS G12D in complex with YK-8S | | Descriptor: | 1-[4-[7-(8-ethynyl-7-fluoranyl-naphthalen-1-yl)-8-fluoranyl-2-[[(2~{R},8~{S})-2-fluoranyl-1,2,3,5,6,7-hexahydropyrrolizin-8-yl]methoxy]pyrido[4,3-d]pyrimidin-4-yl]piperazin-1-yl]-3-oxidanyl-propan-1-one, GTPase KRas, N-terminally processed, ... | | Authors: | Zhang, Z.M, Wang, R.L. | | Deposit date: | 2023-05-23 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.10004044 Å) | | Cite: | Simultaneous Covalent Modification of K-Ras(G12D) and K-Ras(G12C) with Tunable Oxirane Electrophiles.
J.Am.Chem.Soc., 145, 2023
|
|
6V03
 
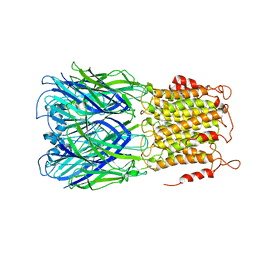 | | ELIC-propylammonium complex in POPC-only nanodiscs | | Descriptor: | 3-AMINOPROPANE, Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Grosman, C, Kumar, P. | | Deposit date: | 2019-11-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of a lipid-sensitive pentameric ligand-gated ion channel embedded in a phosphatidylcholine-only bilayer.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6V0B
 
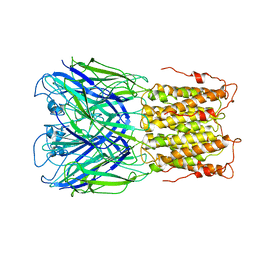 | | Unliganded ELIC in POPC-only nanodiscs. | | Descriptor: | Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Grosman, C, Kumar, P. | | Deposit date: | 2019-11-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of a lipid-sensitive pentameric ligand-gated ion channel embedded in a phosphatidylcholine-only bilayer.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3C6H
 
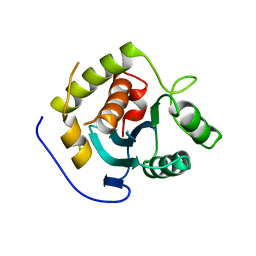 | | Crystal Structure of the RB49 gp17 nuclease domain | | Descriptor: | MAGNESIUM ION, Terminase large subunit | | Authors: | Sun, S, Rossmann, M.G. | | Deposit date: | 2008-02-04 | | Release date: | 2009-01-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces.
Cell(Cambridge,Mass.), 135, 2008
|
|
3CCC
 
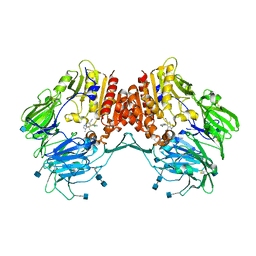 | | Crystal Structure of Human DPP4 in complex with a benzimidazole derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(aminomethyl)-6-(2-chlorophenyl)-1-methyl-1H-benzimidazole-5-carbonitrile, ... | | Authors: | Wallace, M.B, Skene, R.J. | | Deposit date: | 2008-02-25 | | Release date: | 2008-10-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure-based design and synthesis of benzimidazole derivatives as dipeptidyl peptidase IV inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4Y5Q
 
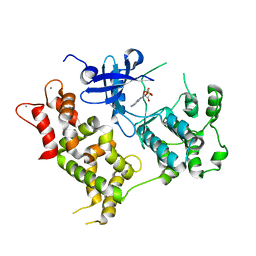 | |
7VJV
 
 | | Human AlkB homolog ALKBH6 in complex with alpha-katoglutarate and Mn | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6, MANGANESE (II) ION | | Authors: | Ma, L, Chen, Z. | | Deposit date: | 2021-09-29 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the interactions and epigenetic functions of human nucleic acid repair protein ALKBH6.
J.Biol.Chem., 298, 2022
|
|
7VJS
 
 | | Human AlkB homolog ALKBH6 in complex with Tris and Ni | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6, NICKEL (II) ION | | Authors: | Ma, L, Chen, Z. | | Deposit date: | 2021-09-28 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural insights into the interactions and epigenetic functions of human nucleic acid repair protein ALKBH6.
J.Biol.Chem., 298, 2022
|
|
8J5X
 
 | | The crystal structure of TrkA(G595R) kinase in complex with N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Descriptor: | High affinity nerve growth factor receptor, N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09192252 Å) | | Cite: | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
8J5W
 
 | | The crystal structure of TrkA(F589L) kinase in complex with N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Descriptor: | High affinity nerve growth factor receptor, N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.28041458 Å) | | Cite: | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
8J61
 
 | | The crystal structure of TrkA kinase in complex with 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Descriptor: | 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide, High affinity nerve growth factor receptor | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05065274 Å) | | Cite: | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
7E3L
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 58G6 heavy chain, 58G6 light chain, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7E3K
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 13G9 heavy chain, 13G9 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
