7SZ4
 
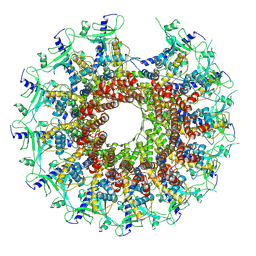 | | Kinetically trapped Pseudomonas-phage PaP3 portal protein - delta barrel mutant class-2 | | Descriptor: | Portal protein | | Authors: | Hou, C.-F.D, Swanson, N.A, Li, F, Yang, R, Lokareddy, R.K, Cingolani, G. | | Deposit date: | 2021-11-25 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM Structure of a Kinetically Trapped Dodecameric Portal Protein from the Pseudomonas-phage PaP3.
J.Mol.Biol., 434, 2022
|
|
7SYA
 
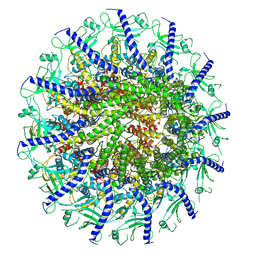 | | Kinetically trapped Pseudomonas-phage PaP3 portal protein - Full Length | | Descriptor: | Portal protein | | Authors: | Hou, C.F.D, Swanson, N.A, Li, F, Yang, R, Lokareddy, R.K, Cingolani, G. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structure of a Kinetically Trapped Dodecameric Portal Protein from the Pseudomonas-phage PaP3.
J.Mol.Biol., 434, 2022
|
|
7SXK
 
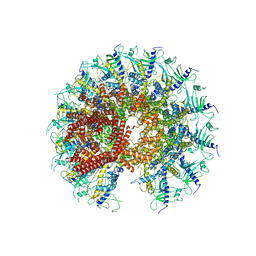 | | Kinetically trapped Pseudomonas-phage PaP3 portal protein - Full Length | | Descriptor: | Portal protein | | Authors: | Hou, C.F.D, Swanson, N.A, Li, F, Yang, R, Lokareddy, R.K, Cingolani, G. | | Deposit date: | 2021-11-23 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structure of a Kinetically Trapped Dodecameric Portal Protein from the Pseudomonas-phage PaP3.
J.Mol.Biol., 434, 2022
|
|
6ZUO
 
 | | Human RIO1(kd)-StHA late pre-40S particle, structural state A (pre 18S rRNA cleavage) | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Plassart, L, Shayan, R, Plisson-Chastang, C. | | Deposit date: | 2020-07-23 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The final step of 40S ribosomal subunit maturation is controlled by a dual key lock.
Elife, 10, 2021
|
|
6ZV6
 
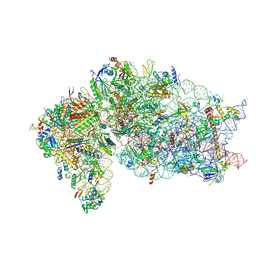 | | Human RIO1(kd)-StHA late pre-40S particle, structural state B (post 18S rRNA cleavage) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Plassart, L, Shayan, R, Plisson-Chastang, C. | | Deposit date: | 2020-07-24 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The final step of 40S ribosomal subunit maturation is controlled by a dual key lock.
Elife, 10, 2021
|
|
1LJE
 
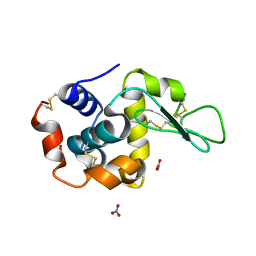 | |
1LJH
 
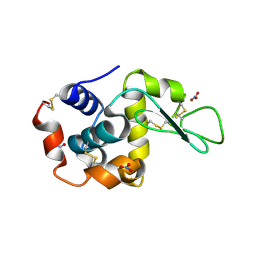 | |
1LJ3
 
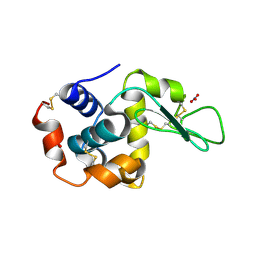 | |
1LJK
 
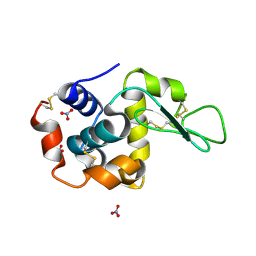 | |
1LJ4
 
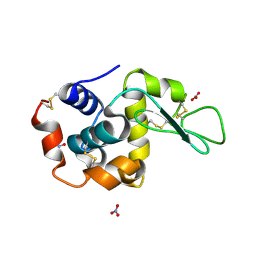 | |
1LJI
 
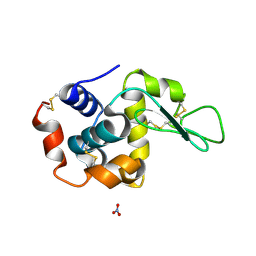 | |
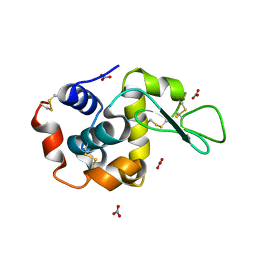
1LJJ
 
 | |
1LJF
 
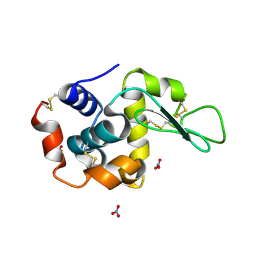 | |
1LJG
 
 | |
1MWN
 
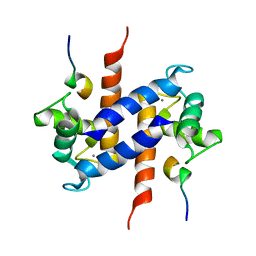 | | Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12 | | Descriptor: | CALCIUM ION, F-actin capping protein alpha-1 subunit, S-100 protein, ... | | Authors: | Inman, K.G, Yang, R, Rustandi, R.R, Miller, K.E, Baldisseri, D.M, Weber, D.J. | | Deposit date: | 2002-09-30 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12
J.Mol.Biol., 324, 2002
|
|
6FVG
 
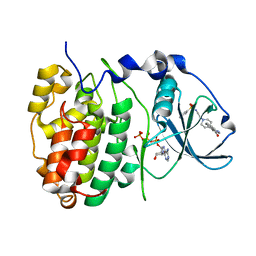 | | The Structure of CK2alpha with CCh507 bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, [1-[2-(phenylsulfonylamino)ethyl]piperidin-4-yl]methyl 1~{H}-indole-3-carboxylate | | Authors: | Brear, P, Prudent, R, Laudet, B, Filhol, O, Cochet, C, Sautel, C, Moucadel, V, Bestgen, B, Engel, M, Ettaoussi, M, Lomberget, T, Le Borgne, M, Kufareva, I, Abagyan, R, Hyvonen, M. | | Deposit date: | 2018-03-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of holoenzyme-disrupting chemicals as substrate-selective CK2 inhibitors.
Sci Rep, 9, 2019
|
|
6FVF
 
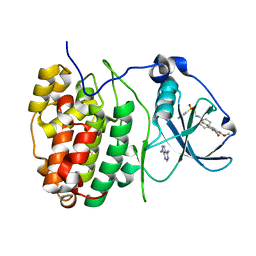 | | The Structure of CK2alpha with CCh503 bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, [1-[2-(phenylsulfonylamino)ethyl]piperidin-4-yl]methyl 5-fluoranyl-2-methoxy-1~{H}-indole-3-carboxylate | | Authors: | Brear, P, Prudent, R, Laudet, B, Filhol, O, Cochet, C, Sautel, C, Moucadel, V, Bestgen, B, Engel, M, Ettaoussi, M, Lomberget, T, Le Borgne, M, Kufareva, I, Abagyan, R, Hyvonen, M. | | Deposit date: | 2018-03-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of holoenzyme-disrupting chemicals as substrate-selective CK2 inhibitors.
Sci Rep, 9, 2019
|
|
3RZE
 
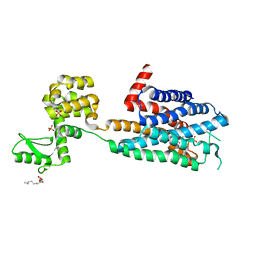 | | Structure of the human histamine H1 receptor in complex with doxepin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (3E)-3-(dibenzo[b,e]oxepin-11(6H)-ylidene)-N,N-dimethylpropan-1-amine, (3Z)-3-(dibenzo[b,e]oxepin-11(6H)-ylidene)-N,N-dimethylpropan-1-amine, ... | | Authors: | Shimamura, T, Han, G.W, Shiroishi, M, Weyand, S, Tsujimoto, H, Winter, G, Katritch, V, Abagyan, R, Cherezov, V, Liu, W, Kobayashi, T, Stevens, R, Iwata, S, GPCR Network (GPCR) | | Deposit date: | 2011-05-11 | | Release date: | 2011-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the human histamine H1 receptor complex with doxepin.
Nature, 475, 2011
|
|
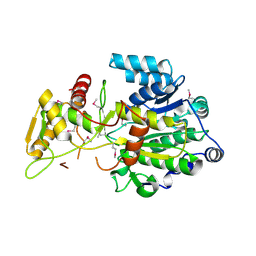
4DQV
 
 | |
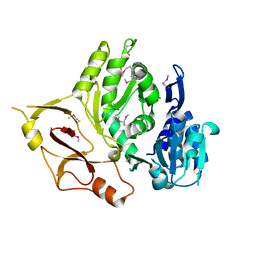
3E53
 
 | |
5T1A
 
 | | Structure of CC Chemokine Receptor 2 with Orthosteric and Allosteric Antagonists | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{R})-1-(4-chloranyl-2-fluoranyl-phenyl)-2-cyclohexyl-3-ethanoyl-4-oxidanyl-2~{H}-pyrrol-5-one, (3S)-1-{(1S,2R,4R)-4-[methyl(propan-2-yl)amino]-2-propylcyclohexyl}-3-{[6-(trifluoromethyl)quinazolin-4-yl]amino}pyrrolidin-2-one, ... | | Authors: | Zheng, Y, Qin, L, Ortiz Zacarias, N.V, de Vries, H, Han, G.W, Gustavsson, M, Dabros, M, Zhao, C, Cherney, R.J, Carter, P, Stamos, D, Abagyan, R, Cherezov, V, Stevens, R.C, IJzerman, A.P, Heitman, L.H, Tebben, A, Kufareva, I, Handel, T.M. | | Deposit date: | 2016-08-18 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists.
Nature, 540, 2016
|
|
5XYH
 
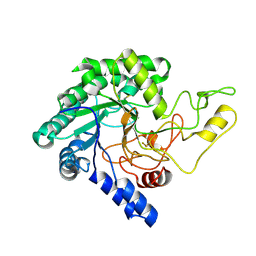 | | Crystal Structure of catalytic domain of 1,4-beta-Cellobiosidase (CbsA) from Xanthomonas oryzae pv. oryzae | | Descriptor: | CbsA | | Authors: | Kumar, S, Haque, A.S, Nathawat, R, Sankaranaryanan, R. | | Deposit date: | 2017-07-07 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | A mutation in an exoglucanase of Xanthomonas oryzae pv. oryzae, which confers an endo mode of activity, affects bacterial virulence, but not the induction of immune responses, in rice
Mol. Plant Pathol., 19, 2018
|
|
4RR7
 
 | | N-terminal editing domain of threonyl-tRNA synthetase from Aeropyrum pernix with L-Ser3AA (snapshot 2) | | Descriptor: | MAGNESIUM ION, Probable threonine--tRNA ligase 2, SERINE-3'-AMINOADENOSINE | | Authors: | Ahmad, S, Muthukumar, S, Yerabham, A.S.K, Kamarthapu, V, Sankaranarayanan, R. | | Deposit date: | 2014-11-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Specificity and catalysis hardwired at the RNA-protein interface in a translational proofreading enzyme.
Nat Commun, 6, 2015
|
|
4RRG
 
 | | Editing domain of threonyl-tRNA synthetase from Methanococcus jannaschii with L-Thr3AA | | Descriptor: | 3'-deoxy-3'-(L-threonylamino)adenosine, Threonine--tRNA ligase | | Authors: | Ahmad, S, Yerabham, A.S.K, Kamarthapu, V, Sankaranarayanan, R. | | Deposit date: | 2014-11-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Specificity and catalysis hardwired at the RNA-protein interface in a translational proofreading enzyme.
Nat Commun, 6, 2015
|
|
4RRK
 
 | |
