2CCK
 
 | | CRYSTAL STRUCTURE OF UNLIGANDED S. AUREUS THYMIDYLATE KINASE | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THYMIDYLATE KINASE | | Authors: | Kotaka, M, Dhaliwal, B, Ren, J, Nichols, C.E, Angell, R, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-01-16 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of S. Aureus Thymidylate Kinase Reveal an Atypical Active Site Configuration and an Intermediate Conformational State Upon Substrate Binding
Protein Sci., 15, 2006
|
|
2CCJ
 
 | | Crystal structure of S. aureus thymidylate kinase complexed with thymidine monophosphate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THYMIDINE-5'-PHOSPHATE, ... | | Authors: | Kotaka, M, Dhaliwal, B, Ren, J, Nichols, C.E, Angell, R, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-01-16 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of S. Aureus Thymidylate Kinase Reveal an Atypical Active Site Configuration and an Intermediate Conformational State Upon Substrate Binding
Protein Sci., 15, 2006
|
|
2C5U
 
 | | T4 RNA Ligase (Rnl1) Crystal Structure | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | El Omari, K, Ren, J, Bird, L.E, Bona, M.K, Klarmann, G, LeGrice, S.F.J, Stammers, D.K. | | Deposit date: | 2005-11-01 | | Release date: | 2005-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Molecular Architecture and Ligand Recognition Determinants for T4 RNA Ligase
J.Biol.Chem., 281, 2006
|
|
1XAI
 
 | | CRYSTAL STRUCTURE OF STAPHLYOCOCCUS AUREUS 3-DEHYDROQUINATE SYNTHASE (DHQS) IN COMPLEX WITH ZN2+, NAD+ AND CARBAPHOSPHONATE | | Descriptor: | 3-dehydroquinate synthase, ZINC ION, [1R-(1ALPHA,3BETA,4ALPHA,5BETA)]-5-(PHOSPHONOMETHYL)-1,3,4-TRIHYDROXYCYCLOHEXANE-1-CARBOXYLIC ACID | | Authors: | Nichols, C.E, Ren, J, Leslie, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-25 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparison of ligand induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases.
J.Mol.Biol., 343, 2004
|
|
3DOL
 
 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with GW695634. | | Descriptor: | N-({4-[({4-chloro-2-[(3-chloro-5-cyanophenyl)carbonyl]phenoxy}acetyl)amino]-3-methylphenyl}sulfonyl)propanamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Chamberlain, P.P, Ren, J, Stammers, D.K. | | Deposit date: | 2008-07-04 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3DOK
 
 | | Crystal structure of K103N mutant HIV-1 reverse transcriptase in complex with GW678248. | | Descriptor: | 2-{4-chloro-2-[(3-chloro-5-cyanophenyl)carbonyl]phenoxy}-N-(2-methyl-4-sulfamoylphenyl)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Chamberlain, P.P, Ren, J, Stammers, D.K. | | Deposit date: | 2008-07-04 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3DMJ
 
 | | CRYSTAL STRUCTURE OF HIV-1 V106A and Y181C MUTANT REVERSE TRANSCRIPTASE IN COMPLEX WITH GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-07-01 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3DLE
 
 | |
3DM2
 
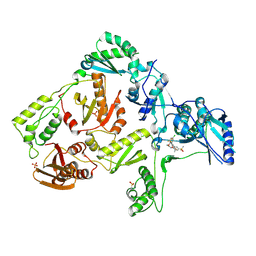 | | Crystal structure of HIV-1 K103N mutant reverse transcriptase in complex with GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3G3Z
 
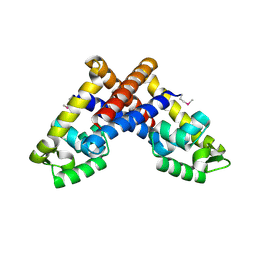 | | The structure of NMB1585, a MarR family regulator from Neisseria meningitidis | | Descriptor: | Transcriptional regulator, MarR family | | Authors: | Nichols, C.E, Sainsbury, S, Ren, J, Walter, T.S, Verma, A, Stammers, D.K, Saunders, N.J, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-02-03 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of NMB1585, a MarR-family regulator from Neisseria meningitidis
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1XAL
 
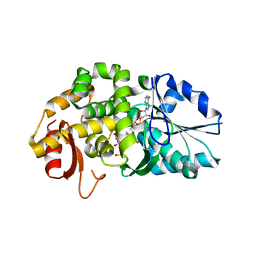 | | CRYSTAL STRUCTURE OF STAPHLYOCOCCUS AUREUS 3-DEHYDROQUINATE SYNTHASE (DHQS) IN COMPLEX WITH ZN2+, NAD+ AND CARBAPHOSPHONATE (SOAK) | | Descriptor: | 3-dehydroquinate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION, ... | | Authors: | Nichols, C.E, Ren, J, Leslie, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-26 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparison of ligand induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases.
J.Mol.Biol., 343, 2004
|
|
1XAG
 
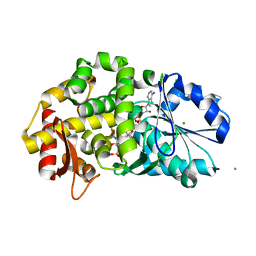 | | CRYSTAL STRUCTURE OF STAPHLYOCOCCUS AUREUS 3-DEHYDROQUINATE SYNTHASE (DHQS) IN COMPLEX WITH ZN2+, NAD+ AND CARBAPHOSPHONATE | | Descriptor: | 3-dehydroquinate synthase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Nichols, C.E, Ren, J, Leslie, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-25 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Comparison of ligand induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases.
J.Mol.Biol., 343, 2004
|
|
1XAH
 
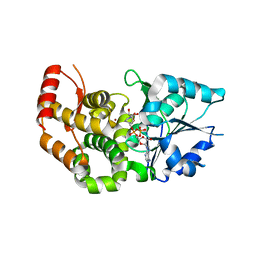 | | CRYSTAL STRUCTURE OF STAPHLYOCOCCUS AUREUS 3-DEHYDROQUINATE SYNTHASE (DHQS) IN COMPLEX WITH ZN2+ AND NAD+ | | Descriptor: | 3-dehydroquinate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Nichols, C.E, Ren, J, Leslie, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-25 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparison of ligand induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases.
J.Mol.Biol., 343, 2004
|
|
1AJ0
 
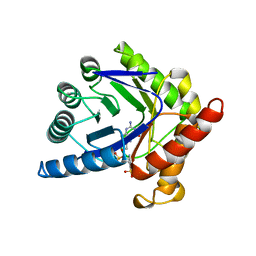 | | CRYSTAL STRUCTURE OF A TERNARY COMPLEX OF E. COLI DIHYDROPTEROATE SYNTHASE | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, DIHYDROPTEROATE SYNTHASE, SULFANILAMIDE, ... | | Authors: | Achari, A, Somers, D.O, Champness, J.N, Bryant, P.K, Rosemond, J, Stammers, D.K. | | Deposit date: | 1997-05-14 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase.
Nat.Struct.Biol., 4, 1997
|
|
1AJ2
 
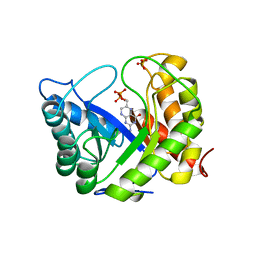 | | CRYSTAL STRUCTURE OF A BINARY COMPLEX OF E. COLI DIHYDROPTEROATE SYNTHASE | | Descriptor: | DIHYDROPTEROATE SYNTHASE, SULFATE ION, [7,8-DIHYDRO-PTERIN-6-YL METHANYL]-PHOSPHONOPHOSPHATE | | Authors: | Achari, A, Somers, D.O, Champness, J.N, Bryant, P.K, Rosemond, J, Stammers, D.K. | | Deposit date: | 1997-05-14 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase.
Nat.Struct.Biol., 4, 1997
|
|
1AJZ
 
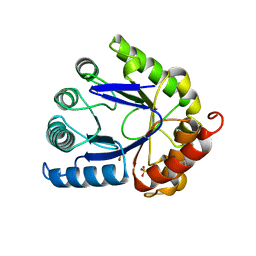 | | STRUCTURE OF DIHYDROPTEROATE PYROPHOSPHORYLASE | | Descriptor: | DIHYDROPTEROATE SYNTHASE, SULFATE ION | | Authors: | Achari, A, Somers, D.O, Champness, J.N, Bryant, P.K, Rosemond, J, Stammers, D.K. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase.
Nat.Struct.Biol., 4, 1997
|
|
2GW8
 
 | | Structure of the PII signal transduction protein of Neisseria meningitidis at 1.85 resolution | | Descriptor: | PII signal transduction protein | | Authors: | Nichols, C.E, Sainsbury, S, Berrow, N.S, Alderton, D, Stammers, D.K, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2006-05-04 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the P(II) signal transduction protein of Neisseria meningitidis at 1.85 A resolution.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2J0W
 
 | | Crystal structure of E. coli aspartokinase III in complex with aspartate and ADP (R-state) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ASPARTIC ACID, CHLORIDE ION, ... | | Authors: | Kotaka, M, Ren, J, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-08-07 | | Release date: | 2006-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of R- and T-State Escherichia Coli Aspartokinase III: Mechanisms of the Allosteric Transition and Inhibition by Lysine.
J.Biol.Chem., 281, 2006
|
|
2J41
 
 | | Crystal structure of Staphylococcus aureus guanylate monophosphate kinase | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, GUANYLATE KINASE, POTASSIUM ION, ... | | Authors: | El Omari, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-08-24 | | Release date: | 2006-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Staphylococcus Aureus Guanylate Monophosphate Kinase
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2GEM
 
 | |
2J0F
 
 | | Structural basis for non-competitive product inhibition in human thymidine phosphorylase: implication for drug design | | Descriptor: | THYMIDINE PHOSPHORYLASE, THYMINE | | Authors: | El Omari, K, Bronckaers, A, Liekens, S, Perez-Perez, M.J, Balzarini, J, Stammers, D.K. | | Deposit date: | 2006-08-02 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis for Non-Competitive Product Inhibition in Human Thymidine Phosphorylase: Implications for Drug Design.
Biochem.J., 399, 2006
|
|
2J87
 
 | | Structure of vaccinia virus thymidine kinase in complex with dTTP: insights for drug design | | Descriptor: | MAGNESIUM ION, THYMIDINE KINASE, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | El Omari, K, Solaroli, N, Karlsson, A, Balzarini, J, Stammers, D.K. | | Deposit date: | 2006-10-23 | | Release date: | 2006-11-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Vaccinia Virus Thymidine Kinase in Complex with Dttp: Insights for Drug Design.
Bmc Struct.Biol., 6, 2006
|
|
2J0X
 
 | | CRYSTAL STRUCTURE OF E. COLI ASPARTOKINASE III IN COMPLEX WITH LYSINE AND ASPARTATE (T-STATE) | | Descriptor: | ASPARTIC ACID, LYSINE, LYSINE-SENSITIVE ASPARTOKINASE 3, ... | | Authors: | Kotaka, M, Ren, J, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-08-07 | | Release date: | 2006-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of R- and T-State Escherichia Coli Aspartokinase III: Mechanisms of the Allosteric Transition and Inhibition by Lysine.
J.Biol.Chem., 281, 2006
|
|
2GEL
 
 | |
2HND
 
 | | Crystal Structure of K101E Mutant HIV-1 Reverse Transcriptase in Complex with Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-12 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
