2PQI
 
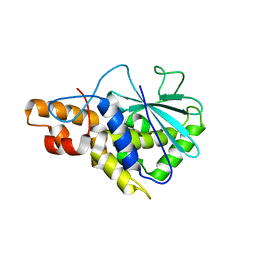 | | Crystal structure of active ribosome inactivating protein from maize (b-32) | | Descriptor: | Ribosome-inactivating protein 3 | | Authors: | Mak, A.N.S, Wong, Y.T, Young, J.A, Cha, S.S, Sze, K.H, Au, S.W.N, Wong, K.B, Shaw, P.C. | | Deposit date: | 2007-05-02 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function study of maize ribosome-inactivating protein: implications for the internal inactivation region and the sole glutamate in the active site.
Nucleic Acids Res., 35, 2007
|
|
2PQG
 
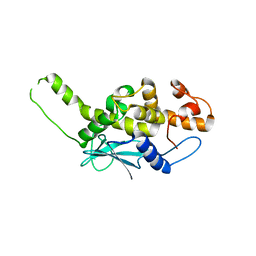 | | Crystal structure of inactive ribosome inactivating protein from maize (b-32) | | Descriptor: | Ribosome-inactivating protein 3 | | Authors: | Mak, A.N.S, Wong, Y.T, Young, J.A, Cha, S.S, Sze, K.H, Au, S.W.N, Wong, K.B, Shaw, P.C. | | Deposit date: | 2007-05-02 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure-function study of maize ribosome-inactivating protein: implications for the internal inactivation region and the sole glutamate in the active site.
Nucleic Acids Res., 35, 2007
|
|
1B9R
 
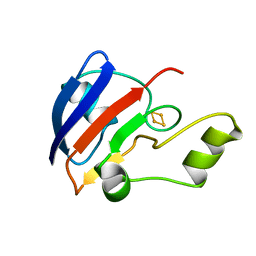 | | TERPREDOXIN FROM PSEUDOMONAS SP. | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, PROTEIN (TERPREDOXIN) | | Authors: | Mo, H, Pochapsky, S.S, Pochapsky, T.C. | | Deposit date: | 1999-02-15 | | Release date: | 1999-05-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A model for the solution structure of oxidized terpredoxin, a Fe2S2 ferredoxin from Pseudomonas.
Biochemistry, 38, 1999
|
|
1ZRR
 
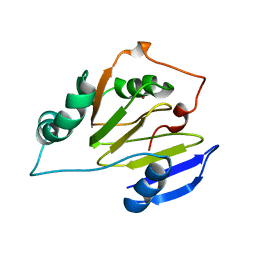 | | Residual Dipolar Coupling Refinement of Acireductone Dioxygenase from Klebsiella | | Descriptor: | E-2/E-2' protein, NICKEL (II) ION | | Authors: | Pochapsky, T.C, Pochapsky, S.S, Ju, T, Hoefler, C, Liang, J. | | Deposit date: | 2005-05-19 | | Release date: | 2005-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A refined model for the structure of acireductone dioxygenase from Klebsiella ATCC 8724 incorporating residual dipolar couplings
J.Biomol.NMR, 34, 2006
|
|
3ISW
 
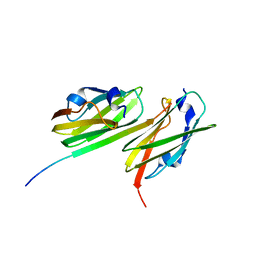 | | Crystal structure of filamin-A immunoglobulin-like repeat 21 bound to an N-terminal peptide of CFTR | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, Filamin-A | | Authors: | Xu, Z, Page, R, Qin, J, Ithychanda, S.S, Liu, J.M, Misra, S. | | Deposit date: | 2009-08-27 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical basis of the interaction between cystic fibrosis transmembrane conductance regulator and immunoglobulin-like repeats of filamin.
J.Biol.Chem., 285, 2010
|
|
2LQD
 
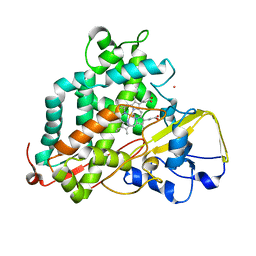 | | Reduced and CO-bound cytochrome P450cam (CYP101A1) | | Descriptor: | CARBON MONOXIDE, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Pochapsky, T.C, Pochapsky, S.S, Asciutto, E, Madura, J, Young, M.J. | | Deposit date: | 2012-03-02 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structural Ensembles of Substrate-Free Cytochrome P450(cam).
Biochemistry, 51, 2012
|
|
5TSW
 
 | |
1KVO
 
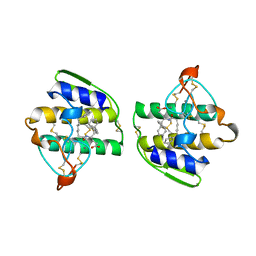 | | HUMAN PHOSPHOLIPASE A2 COMPLEXED WITH A HIGHLY POTENT SUBSTRATE ANOLOGUE | | Descriptor: | 4-(S)-[(1-OXO-7-PHENYLHEPTYL)AMINO]-5-[4-(PHENYLMETHYL)PHENYLTHIO]PENTANOIC ACID, CALCIUM ION, HUMAN PHOSPHOLIPASE A2 | | Authors: | Cha, S.-S, Abdel-Meguid, S.S, Oh, B.-H. | | Deposit date: | 1996-07-29 | | Release date: | 1997-07-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution X-ray crystallography reveals precise binding interactions between human nonpancreatic secreted phospholipase A2 and a highly potent inhibitor (FPL67047XX).
J.Med.Chem., 39, 1996
|
|
4TSV
 
 | |
4IVK
 
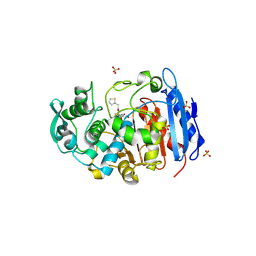 | | Crystal structure of a fammily VIII carboxylesterase in a complex with cephalothin. | | Descriptor: | CEPHALOTHIN GROUP, Carboxylesterases, SULFATE ION | | Authors: | An, Y.J, Kim, M.-K, Jeong, C.-S, Cha, S.-S. | | Deposit date: | 2013-01-23 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the beta-lactamase activity of EstU1, a family VIII carboxylesterase.
Proteins, 81, 2013
|
|
4IVI
 
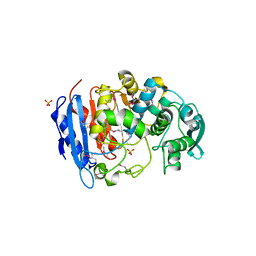 | | Crystal structure of a family VIII carboxylesterase. | | Descriptor: | Carboxylesterase, SULFATE ION | | Authors: | An, Y.J, Kim, M.-K, Jeong, C.-S, Cha, S.-S. | | Deposit date: | 2013-01-23 | | Release date: | 2013-06-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the beta-lactamase activity of EstU1, a family VIII carboxylesterase.
Proteins, 81, 2013
|
|
7RRG
 
 | |
1D2Q
 
 | | CRYSTAL STRUCTURE OF HUMAN TRAIL | | Descriptor: | TNF-RELATED APOPTOSIS INDUCING LIGAND | | Authors: | Cha, S.-S. | | Deposit date: | 1999-09-27 | | Release date: | 2000-02-11 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8 A resolution crystal structure of human TRAIL, a cytokine with selective antitumor activity.
Immunity, 11, 1999
|
|
7L1B
 
 | |
7L1D
 
 | |
7L1C
 
 | |
2K9U
 
 | |
1W02
 
 | |
1W00
 
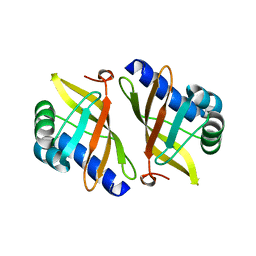 | | Crystal structure of mutant enzyme D103L of Ketosteroid Isomerase from Pseudomonas putida biotype B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 2004-05-30 | | Release date: | 2005-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Double-Mutant Cycle Analysis of a Hydrogen Bond Network in Ketosteroid Isomerase from Pseudomonas Putida Biotype B
Biochem.J., 382, 2004
|
|
5GL4
 
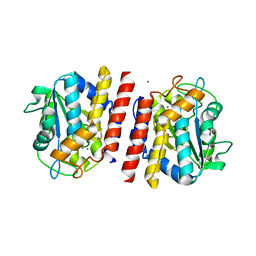 | | Crystal structure of TON_0340 in complex with Mn | | Descriptor: | MANGANESE (II) ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-08 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
5GKX
 
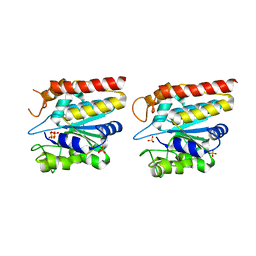 | | Crystal structure of TON_0340, apo form | | Descriptor: | PHOSPHATE ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
4OOZ
 
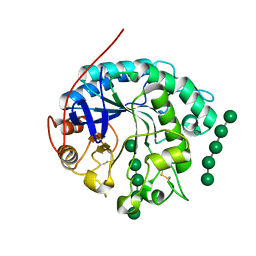 | | Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus in complex with mannopentaose | | Descriptor: | Beta-1,4-mannanase, beta-D-mannopyranose, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, ... | | Authors: | Kim, M.-K, An, Y.J, Jeong, C.-S, Cha, S.-S. | | Deposit date: | 2014-02-04 | | Release date: | 2014-08-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based investigation into the functional roles of the extended loop and substrate-recognition sites in an endo-beta-1,4-d-mannanase from the Antarctic springtail, Cryptopygus antarcticus.
Proteins, 82, 2014
|
|
4OOU
 
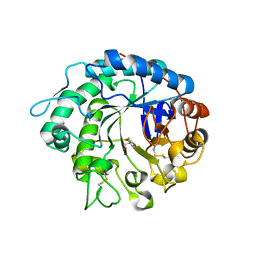 | | Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-1,4-mannanase | | Authors: | Kim, M.-K, An, Y.J, Jeong, C.-S, Cha, S.-S. | | Deposit date: | 2014-02-04 | | Release date: | 2014-08-06 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-based investigation into the functional roles of the extended loop and substrate-recognition sites in an endo-beta-1,4-d-mannanase from the Antarctic springtail, Cryptopygus antarcticus.
Proteins, 82, 2014
|
|
7CS1
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-Complex with Coenzyme A and Neomycin | | Descriptor: | Aminoglycoside 2'-N-acetyltransferase, COENZYME A, NEOMYCIN | | Authors: | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.966 Å) | | Cite: | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
7CSI
 
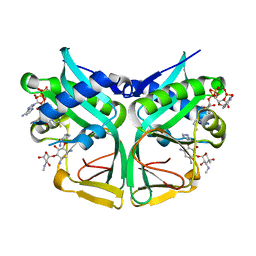 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-Complex with Coenzyme A and Sisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, Aminoglycoside 2'-N-acetyltransferase, COENZYME A | | Authors: | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
