1OZB
 
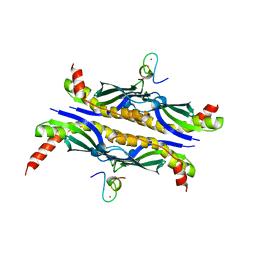 | | Crystal Structure of SecB complexed with SecA C-terminus | | Descriptor: | Preprotein translocase secA subunit, Protein-export protein secB, ZINC ION | | Authors: | Zhou, J, Xu, Z. | | Deposit date: | 2003-04-08 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural determinants of SecB recognition by SecA in bacterial protein translocation
NAT.STRUCT.BIOL., 10, 2003
|
|
1NW1
 
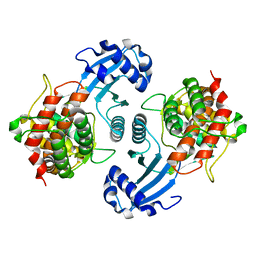 | | Crystal Structure of Choline Kinase | | Descriptor: | CALCIUM ION, Choline kinase (49.2 kD) | | Authors: | Peisach, D, Gee, P, Kent, C, Xu, Z. | | Deposit date: | 2003-02-05 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Crystal Structure of Choline Kinase Reveals a Eukaryotic Protein Kinase Fold
Structure, 11, 2003
|
|
1YTV
 
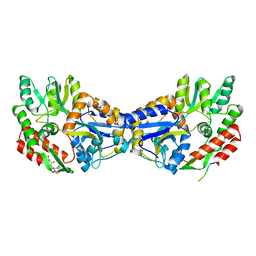 | | Maltose-binding protein fusion to a C-terminal fragment of the V1a vasopressin receptor | | Descriptor: | Maltose-binding periplasmic protein, Vasopressin V1a receptor, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Adikesavan, N.V, Mahmood, S.S, Stanley, S, Xu, Z, Wu, N, Thibonnier, M, Shoham, M. | | Deposit date: | 2005-02-11 | | Release date: | 2005-04-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A C-terminal segment of the V1R vasopressin receptor is unstructured in the crystal structure of its chimera with the maltose-binding protein.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1ZCP
 
 | |
1UKF
 
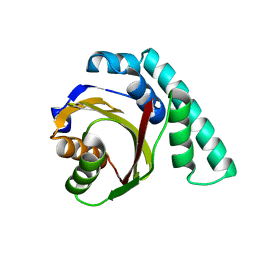 | | Crystal Structure of Pseudomonas Avirulence Protein AvrPphB | | Descriptor: | Avirulence protein AVRPPH3 | | Authors: | Zhu, M, Shao, F, Innes, R.W, Dixon, J.E, Xu, Z. | | Deposit date: | 2003-08-21 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The crystal structure of Pseudomonas avirulence protein AvrPphB: a papain-like fold with a distinct substrate-binding site.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1R62
 
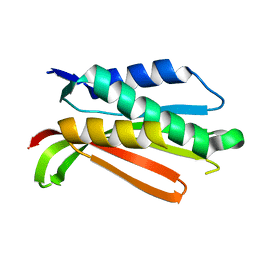 | | Crystal structure of the C-terminal Domain of the Two-Component System Transmitter Protein NRII (NtrB) | | Descriptor: | Nitrogen regulation protein NR(II) | | Authors: | Song, Y, Peisach, D, Pioszak, A.A, Xu, Z, Ninfa, A.J. | | Deposit date: | 2003-10-14 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the C-terminal Domain of the Two-Component System Transmitter Protein Nitrogen Regulator II (NRII; NtrB), Regulator of Nitrogen Assimilation in Escherichia coli.
Biochemistry, 43, 2004
|
|
1T11
 
 | | Trigger Factor | | Descriptor: | Trigger factor | | Authors: | Ludlam, A.V, Moore, B.A, Xu, Z. | | Deposit date: | 2004-04-14 | | Release date: | 2004-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of ribosomal chaperone trigger factor from Vibrio cholerae.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2HZ6
 
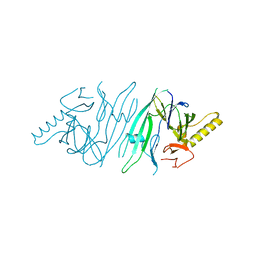 | | The crystal structure of human IRE1-alpha luminal domain | | Descriptor: | Endoplasmic reticulum to nucleus signalling 1 isoform 1 variant | | Authors: | Kaufman, R.J, Xu, Z, Zhou, J. | | Deposit date: | 2006-08-08 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2LP0
 
 | |
3DYR
 
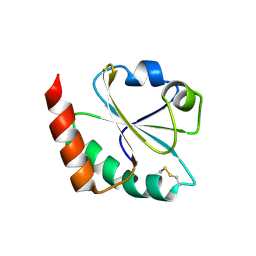 | |
3GGZ
 
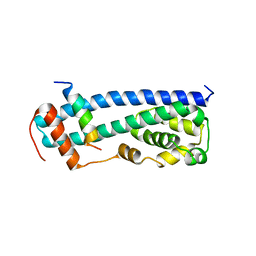 | |
3GGY
 
 | | Crystal Structure of S.cerevisiae Ist1 N-terminal domain | | Descriptor: | Increased sodium tolerance protein 1 | | Authors: | Xiao, J, Xu, Z. | | Deposit date: | 2009-03-02 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis.
MOLECULAR BIOLOGY OF THE CELL, 20, 2009
|
|
6MMZ
 
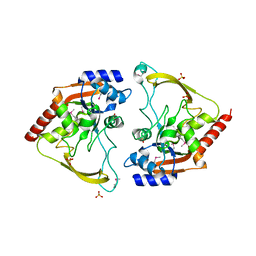 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, H29A mutant apoenzyme | | Descriptor: | Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, SULFATE ION | | Authors: | Stogios, P.J, Skarina, T, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
2LKO
 
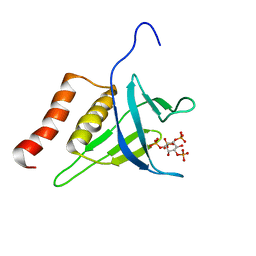 | | Structural Basis of Phosphoinositide Binding to Kindlin-2 Pleckstrin Homology Domain in Regulating Integrin Activation | | Descriptor: | Fermitin family homolog 2, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE | | Authors: | Liu, J, Fukuda, K, Xu, Z. | | Deposit date: | 2011-10-17 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of phosphoinositide binding to kindlin-2 protein pleckstrin homology domain in regulating integrin activation.
J.Biol.Chem., 286, 2011
|
|
4GVD
 
 | | Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PDZ in complex with Syndecan1 Peptide | | Descriptor: | 5-(DIMETHYLAMINO)-1-NAPHTHALENESULFONIC ACID(DANSYL ACID), CHLORIDE ION, SODIUM ION, ... | | Authors: | Liu, X, Shepherd, T.R, Murray, A.M, Xu, Z, Fuentes, E.J. | | Deposit date: | 2012-08-30 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of the Tiam1 PDZ domain/ phospho-syndecan1 complex reveals a ligand conformation that modulates protein dynamics.
Structure, 21, 2013
|
|
4GVC
 
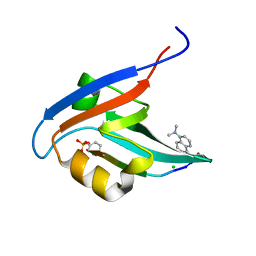 | | Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PDZ in complex with phosphorylated Syndecan1 Peptide | | Descriptor: | 5-(DIMETHYLAMINO)-1-NAPHTHALENESULFONIC ACID(DANSYL ACID), CHLORIDE ION, SODIUM ION, ... | | Authors: | Liu, X, Shepherd, T.R, Murray, A.M, Xu, Z, Fuentes, E.J. | | Deposit date: | 2012-08-30 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The structure of the Tiam1 PDZ domain/ phospho-syndecan1 complex reveals a ligand conformation that modulates protein dynamics.
Structure, 21, 2013
|
|
2OCG
 
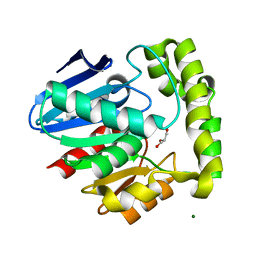 | | Crystal structure of human valacyclovir hydrolase | | Descriptor: | GLYCEROL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Lai, L, Xu, Z, Amidon, G.L. | | Deposit date: | 2006-12-20 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular basis of prodrug activation by human valacyclovirase, an alpha-amino acid ester hydrolase.
J.Biol.Chem., 283, 2008
|
|
2OCI
 
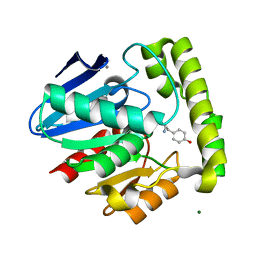 | | Crystal structure of valacyclovir hydrolase complexed with a product analogue | | Descriptor: | L-TYROSINAMIDE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Lai, L, Xu, Z, Amidon, G.L. | | Deposit date: | 2006-12-20 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of prodrug activation by human valacyclovirase, an alpha-amino acid ester hydrolase.
J.Biol.Chem., 283, 2008
|
|
2OCK
 
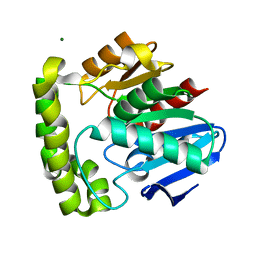 | | Crystal structure of valacyclovir hydrolase D123N mutant | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Valacyclovir hydrolase | | Authors: | Lai, L, Xu, Z, Amidon, G.L. | | Deposit date: | 2006-12-20 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular basis of prodrug activation by human valacyclovirase, an alpha-amino acid ester hydrolase.
J.Biol.Chem., 283, 2008
|
|
2OCL
 
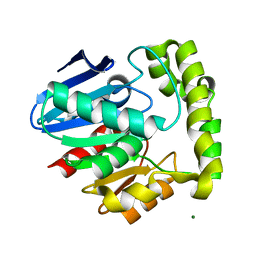 | | Crystal structure of valacyclovir hydrolase S122A mutant | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Valacyclovir hydrolase | | Authors: | Lai, L, Xu, Z, Amidon, G.L. | | Deposit date: | 2006-12-20 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of prodrug activation by human valacyclovirase, an alpha-amino acid ester hydrolase.
J.Biol.Chem., 283, 2008
|
|
2PFV
 
 | |
2PFT
 
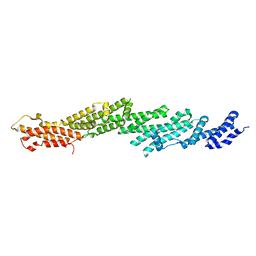 | |
2QP9
 
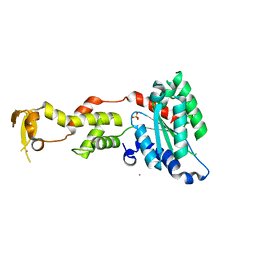 | | Crystal Structure of S.cerevisiae Vps4 | | Descriptor: | CADMIUM ION, SULFATE ION, Vacuolar protein sorting-associated protein 4 | | Authors: | Xiao, J, Xu, Z. | | Deposit date: | 2007-07-23 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural characterization of the ATPase reaction cycle of endosomal AAA protein Vps4.
J.Mol.Biol., 374, 2007
|
|
3LM1
 
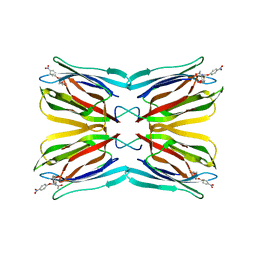 | | Crystal Structure Analysis of Maclura pomifera agglutinin complex with p-nitrophenyl-GalNAc | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Agglutinin alpha chain, Agglutinin beta-2 chain | | Authors: | Huang, J, Xu, Z, Wang, D, Ogato, C, Hirama, T, Palczewski, K, Hazen, S.L, Lee, X, Young, N.M. | | Deposit date: | 2010-01-29 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the secondary binding sites of Maclura pomifera agglutinin by glycan array and crystallographic analyses.
Glycobiology, 20, 2010
|
|
5XS2
 
 | |
