5E1D
 
 | | NTMT1 in complex with YPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
5E1O
 
 | | Crystal structure of NTMT1 in complex with RPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
8P9F
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB161 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-4-[2-(2-azanyl-5-methyl-4-oxidanyl-phenyl)hydrazinyl]benzamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8P9J
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB500 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-5-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)pyridine-2-carboxamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8P9L
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB512 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-azanyl-5-phenyl-phenyl)-5-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)pyridine-2-carboxamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
5DF6
 
 | | Crystal structure of PTPN11 tandem SH2 domains in complex with a TXNIP peptide | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11, UNKNOWN ATOM OR ION, txnip | | Authors: | Dong, A, Li, W, Tempel, W, Liu, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-08-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis for the regulatory role of the PPxY motifs in the thioredoxin-interacting protein TXNIP.
Biochem.J., 473, 2016
|
|
7BBO
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P212121 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212
To Be Published
|
|
7BBP
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with H4K5acK8ac | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Histone H4, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with H4K5acK8ac
To Be Published
|
|
5E1B
 
 | | Crystal structure of NRMT1 in complex with SPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
4Z0O
 
 | | CW-type zinc finger of ZCWPW2 with F78D mutation | | Descriptor: | SODIUM ION, UNKNOWN ATOM OR ION, ZINC ION, ... | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | CW-type zinc finger of ZCWPW2 with F78D mutation
To be Published
|
|
7B9E
 
 | | Crystal structure of MurE from E.coli in complex with Z275151340 | | Descriptor: | 4-chloro-N-cyclopentyl-1-methyl-1H-pyrazole-3-carboxamide, ISOPROPYL ALCOHOL, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-14 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7A6U
 
 | | Cryo-EM structure of the cytoplasmic domain of human TRPC6 | | Descriptor: | Short transient receptor potential channel 6, UNKNOWN ATOM OR ION | | Authors: | Grieben, M, Pike, A.C.W, Wang, D, Mukhopadhyay, S.M.M, Chalk, R, Marsden, B.D, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-26 | | Release date: | 2020-09-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM structure of the cytoplasmic domain of human TRPC6
TO BE PUBLISHED
|
|
6MLC
 
 | | PHD6 domain of MLL3 in complex with histone H4 | | Descriptor: | GLYCEROL, Histone H4, Histone-lysine N-methyltransferase 2C, ... | | Authors: | Dong, A, Liu, Y, Qin, S, Lei, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-27 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into trans-histone regulation of H3K4 methylation by unique histone H4 binding of MLL3/4.
Nat Commun, 10, 2019
|
|
7APF
 
 | | Crystal structure of JAK3 in complex with FM601 (compound 10a) | | Descriptor: | 1,2-ETHANEDIOL, 1-phenylurea, 3-[3-(propanoylamino)phenyl]-1~{H}-pyrrolo[2,3-b]pyridine-5-carboxamide, ... | | Authors: | Chaikuad, A, Forster, M, Gehringer, M, Laufer, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-10-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of a Novel Class of Covalent Dual Inhibitors Targeting the Protein Kinases BMX and BTK.
Int J Mol Sci, 21, 2020
|
|
7APG
 
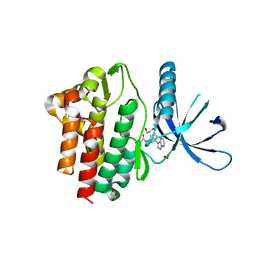 | | Crystal structure of JAK3 in complex with FM587 (compound 9a) | | Descriptor: | 1,2-ETHANEDIOL, 1-phenylurea, Tyrosine-protein kinase JAK3, ... | | Authors: | Chaikuad, A, Forster, M, Gehringer, M, Laufer, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-10-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Novel Class of Covalent Dual Inhibitors Targeting the Protein Kinases BMX and BTK.
Int J Mol Sci, 21, 2020
|
|
7AKG
 
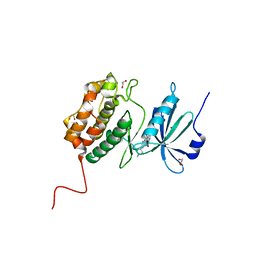 | |
6V9T
 
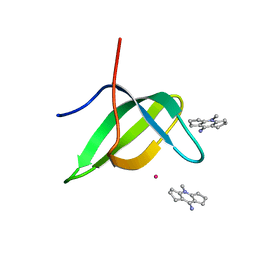 | | Tudor domain of TDRD3 in complex with a small molecule | | Descriptor: | 4-methyl-2,3,4,5,6,7-hexahydrodicyclopenta[b,e]pyridin-8(1H)-imine, Tudor domain-containing protein 3, UNKNOWN ATOM OR ION | | Authors: | Li, W, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-16 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
6Q8P
 
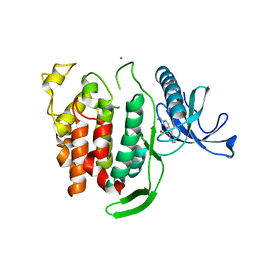 | | Structure of CLK1 with bound N-methyl-10-nitropyrido[3,4-g]quinazolin-2-amine | | Descriptor: | Dual specificity protein kinase CLK1, POTASSIUM ION, ~{N}-methyl-10-nitro-pyrido[3,4-g]quinazolin-2-amine | | Authors: | Joerger, A.C, Chatterjee, D, Schroeder, M, Tazarki, H, Zeinyeh, W, Esvan, Y.J, Khiari, J, Josselin, B, Baratte, B, Bach, S, Ruchaud, S, Anizon, F, Giraud, F, Moreau, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New pyrido[3,4-g]quinazoline derivatives as CLK1 and DYRK1A inhibitors: synthesis, biological evaluation and binding mode analysis.
Eur J Med Chem, 166, 2019
|
|
5FET
 
 | | Crystal Structure of PVX_084705 in presence of Compound 2 | | Descriptor: | 4-[7-[(dimethylamino)methyl]-2-(4-fluorophenyl)imidazo[1,2-a]pyridin-3-yl]pyrimidin-2-amine, cGMP-dependent protein kinase, putative | | Authors: | Wernimont, A.K, Tempel, W, Walker, J.R, He, H, Seitova, A, Hills, T, Neculai, A.M, Baker, D.A, Flueck, C, Kettleborough, C.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, El Bakkouri, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-17 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Crystal Structure of PVX_084705 in presence of Compound 2
To be published
|
|
5FDP
 
 | | Structure of DDR1 receptor tyrosine kinase in complex with D2099 inhibitor at 2.25 Angstroms resolution. | | Descriptor: | (4~{S})-4-methyl-~{N}-[3-[(4-methylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl]-2-pyrimidin-5-yl-3,4-dihydro-1~{H}-isoquinoline-7-carboxamide, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bartual, S.G, Pinkas, D.M, Wang, Z, Ding, K, Mahajan, P, Kupinska, K, Mukhopadhyay, S, Strain-Damerell, C, Borkowska, O, Talon, R, Kopec, J, Williams, E, Tallant, C, Chaikuad, A, Sorell, F, Newman, J, Burgess-Brown, N, Arrowsmith, C.H, von Delft, F, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Design of Tetrahydroisoquinoline-7-carboxamides as Selective Discoidin Domain Receptor 1 (DDR1) Inhibitors.
J.Med.Chem., 59, 2016
|
|
5FDX
 
 | | Structure of DDR1 receptor tyrosine kinase in complex with D2164 inhibitor at 2.65 Angstroms resolution. | | Descriptor: | 1,2-ETHANEDIOL, 3-[(4-methylpiperazin-1-yl)methyl]-~{N}-[(4~{R})-4-methyl-2-pyrimidin-5-yl-3,4-dihydro-1~{H}-isoquinolin-7-yl]-5-(trifluoromethyl)benzamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bartual, S.G, Pinkas, D.M, Wang, Z, Ding, K, Mahajan, P, Kupinska, K, Mukhopadhyay, S, Strain-Damerell, C, Chalk, R, Borkowska, O, Talon, R, Kopec, J, Williams, E, Tallant, C, Chaikuad, A, Sorell, F, Newman, J, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of DDR1 receptor tyrosine kinase in complex with D2164 inhibitor at 2.65 Angstroms resolution.
To Be Published
|
|
8RDK
 
 | | Crystal structure of human Haspin (GSG2) kinase bound to MD420 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-pyridin-4-yl-1~{H}-pyrrolo[3,2-g]isoquinoline, Serine/threonine-protein kinase haspin, ... | | Authors: | Kraemer, A, Defois, M, Giraud, F, Moreau, P, Anizon, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-12-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
7BDO
 
 | | MAPK14 bound with SR302 | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-[[(3~{S})-1-methylsulfonylpiperidin-3-yl]amino]-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Schroeder, M, Roehm, S, Joerger, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
7BDQ
 
 | | MAPK14 bound with SR300 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase 14, ... | | Authors: | Schroeder, M, Roehm, S, Joerger, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
5I3O
 
 | | Crystal Structure of BMP-2-inducible kinase in complex with an Indazole inhibitor | | Descriptor: | BMP-2-inducible protein kinase, N-(6-{3-[(dimethylsulfamoyl)amino]phenyl}-1H-indazol-3-yl)cyclopropanecarboxamide, SULFATE ION | | Authors: | Counago, R.M, Sorrell, F.J, Krojer, T, Savitsky, P, Elkins, J.M, Axtman, A, Drewry, D, Wells, C, Zhang, C, Zuercher, W, Willson, T.M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Arruda, P, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of BMP-2-inducible kinase in complex with an Indazole inhibitor
To Be Published
|
|
