6GWA
 
 | | Concanavalin B structure determined with data from the EuXFEL, the first MHz free electron laser | | Descriptor: | Concanavalin B | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-06-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
4BVX
 
 | | Crystal structure of the AIMP3-MRS N-terminal domain complex with I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, EUKARYOTIC TRANSLATION ELONGATION FACTOR 1 EPSILON-1, METHIONINE--TRNA LIGASE, ... | | Authors: | Cho, H.Y, Seo, W.W, Cho, H.J, Kang, B.S. | | Deposit date: | 2013-06-29 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Assembly of Multi-tRNA Synthetase Complex Via Heterotetrameric Glutathione Transferase-Homology Domains.
J.Biol.Chem., 290, 2015
|
|
3M16
 
 | | Structure of a Transaldolase from Oleispira antarctica | | Descriptor: | Transaldolase | | Authors: | Singer, A.U, Kagan, O, Zhang, R, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-04 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3ZGA
 
 | | Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Carbenicillin bound form | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2S)-2-carboxy-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, PENICILLIN-BINDING PROTEIN 4 | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2012-12-17 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal Structures of Bifunctional Penicillin-Binding Protein 4 from Listeria Monocytogenes.
Antimicrob.Agents Chemother., 57, 2013
|
|
6P5F
 
 | | Photoactive Yellow Protein PYP Pure Dark | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6P5E
 
 | | Photoactive Yellow Protein PYP 80ps | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
3ZPJ
 
 | | Crystal structure of Ton1535 from Thermococcus onnurineus NA1 | | Descriptor: | TON_1535 | | Authors: | Jeong, J.H, kim, Y.G. | | Deposit date: | 2013-02-28 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structure of the Hypothetical Protein Ton1535 from Thermococcus Onnurineus Na1 Reveals Unique Structural Properties by a Left-Handed Helical Turn in Normal Alpha-Solenoid Protein.
Proteins, 82, 2014
|
|
6P5D
 
 | | Photoactive Yellow Protein PYP 30ps | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
7ROA
 
 | | Crystal structure of EntV136 from Enterococcus faecalis | | Descriptor: | EntV | | Authors: | Stogios, P.J, Evdokimova, E, Kim, Y, Garsin, D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2021-07-30 | | Release date: | 2022-10-12 | | Last modified: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural and functional analysis of EntV reveals a 12 amino acid fragment protective against fungal infections.
Nat Commun, 13, 2022
|
|
4BBW
 
 | | The crystal structure of Sialidase VPI 5482 (BTSA) from Bacteroides thetaiotaomicron | | Descriptor: | SIALIDASE (NEURAMINIDASE) | | Authors: | Park, K.-H, Song, H.-N, Jung, T.-Y, Lee, M.-H, Woo, E.-J. | | Deposit date: | 2012-09-28 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Characterization of the Broad Substrate Specificity of Bacteroides Thetaiotaomicron Commensal Sialidase.
Biochim.Biophys.Acta, 1834, 2013
|
|
6P5G
 
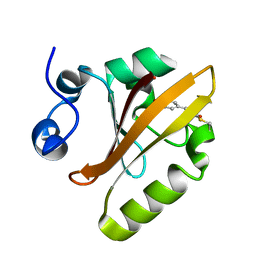 | | Photoactive Yellow Protein PYP Dark Full | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6P4I
 
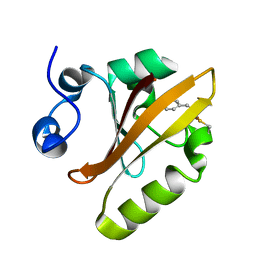 | | Photoactive Yellow Protein PYP 10ps | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-27 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
5DNY
 
 | | Structure of the ATPrS-Mre11/Rad50-DNA complex | | Descriptor: | DNA (27-MER), DNA double-strand break repair Rad50 ATPase,DNA double-strand break repair Rad50 ATPase, DNA double-strand break repair protein Mre11, ... | | Authors: | Liu, Y. | | Deposit date: | 2015-09-10 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex.
Embo J., 35, 2016
|
|
5F3W
 
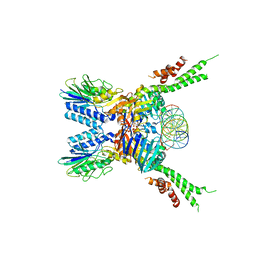 | | Structure of the ATPrS-Mre11/Rad50-DNA complex | | Descriptor: | 27-MER DNA, DNA double-strand break repair Rad50 ATPase,DNA double-strand break repair Rad50 ATPase, DNA double-strand break repair protein Mre11, ... | | Authors: | Liu, Y. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex
Embo J., 35, 2016
|
|
3ZG8
 
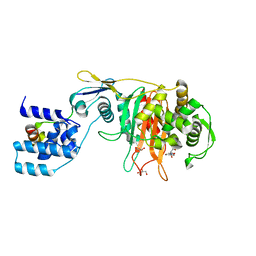 | | Crystal Structure of Penicillin Binding Protein 4 from Listeria monocytogenes in the Ampicillin bound form | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, PENICILLIN-BINDING PROTEIN, ... | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2012-12-17 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Crystal Structures of Bifunctional Penicillin-Binding Protein 4 from Listeria Monocytogenes.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZG7
 
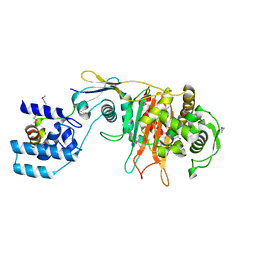 | |
8AXW
 
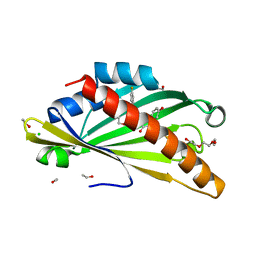 | | The structure of mouse AsterC (GramD1c) with Ezetimibe | | Descriptor: | (3~{R},4~{S})-1-(4-fluorophenyl)-3-[(3~{S})-3-(4-fluorophenyl)-3-oxidanyl-propyl]-4-(4-hydroxyphenyl)azetidin-2-one, CHLORIDE ION, ETHANOL, ... | | Authors: | Fairall, L, Xiao, X, Burger, L, Tontonoz, P, Schwabe, J.W.R. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aster-dependent nonvesicular transport facilitates dietary cholesterol uptake.
Science, 382, 2023
|
|
2QMQ
 
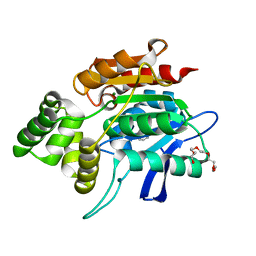 | |
5BS6
 
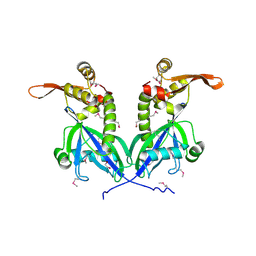 | | Apo structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI | | Descriptor: | 1,2-ETHANEDIOL, transcriptional regulator AraR | | Authors: | Chang, C, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
5A34
 
 | | The crystal structure of the GST-like domains complex of EPRS-AIMP2 | | Descriptor: | AMINOACYL TRNA SYNTHASE COMPLEX-INTERACTING MULTIFUNCTIONAL PROTEIN 2, BIFUNCTIONAL GLUTAMATE/PROLINE--TRNA LIGASE, GLYCEROL | | Authors: | Cho, H.Y, Kang, B.S. | | Deposit date: | 2015-05-27 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Assembly of Multi-tRNA Synthetase Complex Via Heterotetrameric Glutathione Transferase-Homology Domains.
J.Biol.Chem., 290, 2015
|
|
8GUY
 
 | | human insulin receptor bound with two insulin molecules | | Descriptor: | Insulin A chain, Insulin, isoform 2, ... | | Authors: | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
6WEB
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
6WEC
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
8H86
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
8H87
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR2 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR2, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
