5CBU
 
 | | Human Cyclophilin D Complexed with Inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-01 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor.
To Be Published
|
|
5CC9
 
 | |
6H2B
 
 | | Structure of the Macrobrachium rosenbergii Nodavirus | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Ho, K.H, Gabrielsen, M, Beh, P.L, Kueh, C.L, Thong, Q.X, Streetley, J, Tan, W.S, Bhella, D. | | Deposit date: | 2018-07-13 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the Macrobrachium rosenbergii nodavirus: A new genus within the Nodaviridae?
PLoS Biol., 16, 2018
|
|
5C79
 
 | | PH domain of ASAP1 in complex with diC4-PtdIns(4,5)P2 | | Descriptor: | (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1 ,2-DIYL DIBUTANOATE, Arf-GAP, CHLORIDE ION | | Authors: | Xia, D, Tang, W.K. | | Deposit date: | 2015-06-24 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Basis for Cooperative Binding of Anionic Phospholipids to the PH Domain of the Arf GAP ASAP1.
Structure, 23, 2015
|
|
6YLA
 
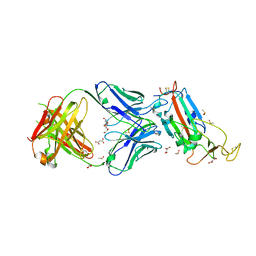 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with CR3022 Fab | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Huo, J, Zhao, Y, Ren, J, Zhou, D, Ginn, H.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-04-06 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6YQ1
 
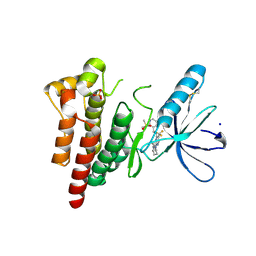 | | FOCAL ADHESION KINASE CATALYTIC DOMAIN IN COMPLEX WITH N-Methyl-N-(3-{[2-(2-oxo-1,2,3,4-tetrahydro-quinolin-6-ylamino)-5-trifluoromethyl-pyrimidin-4-ylamino]-methyl}-pyridin-2-yl)-methanesulfonamide | | Descriptor: | Focal adhesion kinase 1, SODIUM ION, SULFATE ION, ... | | Authors: | Musil, D, Heinrich, T, Amaral, M. | | Deposit date: | 2020-04-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.784 Å) | | Cite: | Structure-kinetic relationship reveals the mechanism of selectivity of FAK inhibitors over PYK2.
Cell Chem Biol, 28, 2021
|
|
6GP3
 
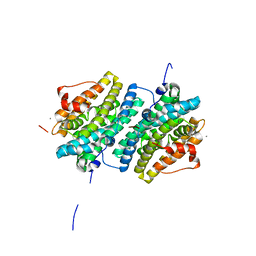 | | Ribonucleotide Reductase class Ie R2 from Mesoplasma florum, inactive form | | Descriptor: | CALCIUM ION, Ribonucleoside-diphosphate reductase beta chain | | Authors: | Srinivas, V, Lebrette, H, Lundin, D, Kutin, Y, Sahlin, M, Lerche, M, Enrich, J, Branca, R.M.M, Cox, N, Sjoberg, B.M, Hogbom, M. | | Deposit date: | 2018-06-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Metal-free ribonucleotide reduction powered by a DOPA radical in Mycoplasma pathogens.
Nature, 563, 2018
|
|
6YR9
 
 | |
6YOJ
 
 | |
1GNU
 
 | | GABA(A) receptor associated protein GABARAP | | Descriptor: | GABARAP, NICKEL (II) ION | | Authors: | Knight, D, Harris, R, Moss, S, Driscoll, P.C, Keep, N.H. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-Ray Crystal Structure and Putative Ligand-Derived Peptide Binding Properties of Gamma-Aminobutyric Acid Receptor Type a Receptor-Associated Protein
J.Biol.Chem., 277, 2002
|
|
6YK8
 
 | | OTU-like deubiquitinase from Legionella -Lpg2529 | | Descriptor: | PLATINUM (II) ION, Uncharacterized protein | | Authors: | Shin, D, Dikic, I. | | Deposit date: | 2020-04-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Bacterial OTU deubiquitinases regulate substrate ubiquitination upon Legionella infection.
Elife, 9, 2020
|
|
6GSH
 
 | | Feline Calicivirus Strain F9 | | Descriptor: | POTASSIUM ION, VP1 | | Authors: | Conley, M.J, Bhella, D. | | Deposit date: | 2018-06-14 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Calicivirus VP2 forms a portal-like assembly following receptor engagement.
Nature, 565, 2019
|
|
5C3H
 
 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 1 | | Descriptor: | 4-[2-oxo-2-(piperidin-1-yl)ethyl]piperazin-1-ium, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
6GTP
 
 | | Structure of the AtaT Y144F mutant toxin | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Garcia-Pino, A, Jurenas, D. | | Deposit date: | 2018-06-18 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of regulation and neutralization of the AtaR-AtaT toxin-antitoxin system.
Nat. Chem. Biol., 15, 2019
|
|
5C3L
 
 | | Structure of the metazoan Nup62.Nup58.Nup54 nucleoporin complex. | | Descriptor: | Nanobody Nb15, Nucleoporin Nup58, Nucleoporin Nup62, ... | | Authors: | Chug, H, Trakhanov, S, Hulsmann, B.B, Pleiner, T, Gorlich, D. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the metazoan Nup62Nup58Nup54 nucleoporin complex.
Science, 350, 2015
|
|
6GVE
 
 | | GAPDH-CP12-PRK complex | | Descriptor: | CP12 polypeptide, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | McFarlane, C.R, Shah, N, Bubeck, D, Murray, J.W. | | Deposit date: | 2018-06-20 | | Release date: | 2019-07-03 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of light-induced redox regulation in the Calvin-Benson cycle in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4I8B
 
 | | Crystal Structure of Thioredoxin from Schistosoma Japonicum | | Descriptor: | Thioredoxin | | Authors: | Wu, Q, Peng, Y, Zhao, J, Li, X, Fan, X, Zhou, X, Chen, J, Luo, Z, Shi, D. | | Deposit date: | 2012-12-03 | | Release date: | 2013-12-04 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Expression, characterization and crystal structure of thioredoxin from Schistosoma japonicum.
Parasitology, 142, 2015
|
|
5C7A
 
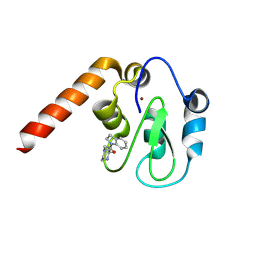 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 7 | | Descriptor: | (2R)-4-[2-(2,3-dihydro-1H-indol-1-yl)-2-oxoethyl]-2-methylpiperazin-1-ium, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
5CBT
 
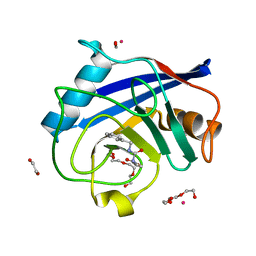 | | Human Cyclophilin D Complexed with Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, POTASSIUM ION, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-01 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor
To Be Published
|
|
6H0R
 
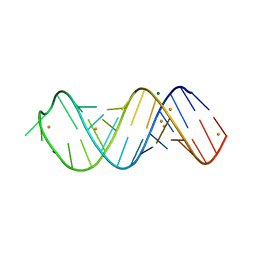 | | X-ray structure of SRS2 fragment of Rgs4 3' UTR | | Descriptor: | BARIUM ION, MAGNESIUM ION, SRS2 fragment of Rgs4 3' UTR, ... | | Authors: | Heber, S, Janowski, R, Niessing, D. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Staufen2-mediated RNA recognition and localization requires combinatorial action of multiple domains.
Nat Commun, 10, 2019
|
|
6H55
 
 | | core of the human pyruvate dehydrogenase (E2) | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | | Authors: | Haselbach, D, Prajapati, S, Tittmann, K, Stark, H. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural and Functional Analyses of the Human PDH Complex Suggest a "Division-of-Labor" Mechanism by Local E1 and E3 Clusters.
Structure, 27, 2019
|
|
6H1T
 
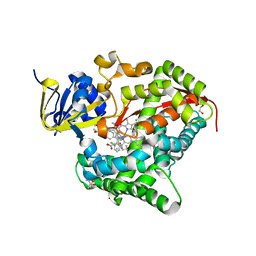 | | Structure of the BM3 heme domain in complex with clotrimazole | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, ... | | Authors: | Jeffreys, L.N, Munro, A.W.M, Leys, D. | | Deposit date: | 2018-07-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Novel insights into P450 BM3 interactions with FDA-approved antifungal azole drugs.
Sci Rep, 9, 2019
|
|
6H5S
 
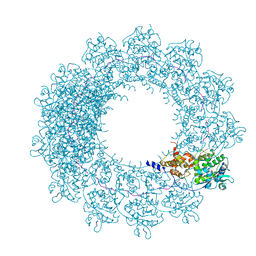 | | Cryo-EM map of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to viral genomic 5-prime RNA hexamers. | | Descriptor: | Nucleocapsid, RNA (5'-R(*AP*CP*CP*AP*GP*A)-3') | | Authors: | Desfosses, A, Milles, S, Ringkjobing Jensen, M, Guseva, S, Colletier, J.P, Maurin, D, Schoehn, G, Gutsche, I, Ruigrok, R, Blackledge, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5BZ3
 
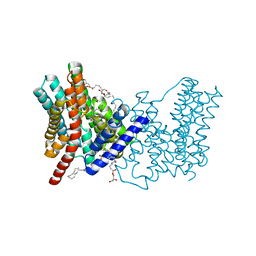 | | CRYSTAL STRUCTURE OF SODIUM PROTON ANTIPORTER NAPA IN OUTWARD-FACING CONFORMATION. | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL (7Z)-TETRADEC-7-ENOATE, Na(+)/H(+) antiporter | | Authors: | Coincon, M, Uzdavinys, P, Emmanuel, N, Cameron, A, Drew, D. | | Deposit date: | 2015-06-11 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures reveal the molecular basis of ion translocation in sodium/proton antiporters.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5C2U
 
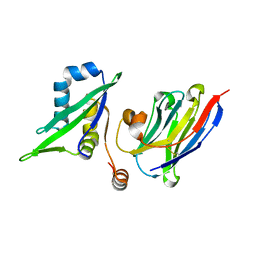 | | Ferredoxin-like domain of nucleoporin Nup54 bound to a nanobody | | Descriptor: | Nanobody, Nup54 | | Authors: | Chug, H, Trakhanov, S, Hulsmann, B.B, Pleiner, T, Gorlich, D. | | Deposit date: | 2015-06-16 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the metazoan Nup62Nup58Nup54 nucleoporin complex.
Science, 350, 2015
|
|
