7CL0
 
 | | Crystal structure of human SIRT6 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | Song, K, Zhang, J. | | Deposit date: | 2020-07-20 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Reply to: Binding site for MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
7CL1
 
 | | Human SIRT6 in complex with allosteric activator MDL-801 (3.2A) | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Song, K, Zhang, J. | | Deposit date: | 2020-07-20 | | Release date: | 2021-02-24 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Reply to: Binding site for MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
1UP7
 
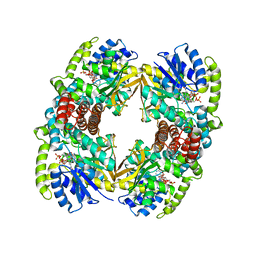 | | Structure of the 6-phospho-beta glucosidase from Thermotoga maritima at 2.4 Angstrom resolution in the tetragonal form with NAD and glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-PHOSPHO-BETA-GLUCOSIDASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Varrot, A, Yip, V.L, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-09-29 | | Release date: | 2004-11-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nad+ and Metal-Ion Dependent Hydrolysis by Family 4 Glycosidases: Structural Insight Into Specificity for Phospho-Beta-D-Glucosides
J.Mol.Biol., 346, 2005
|
|
8ZUF
 
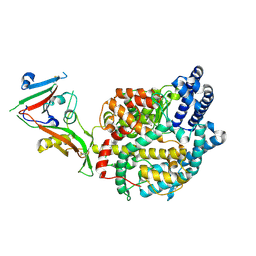 | | Cryo-EM structure of P.nat ACE2 mutant in complex with MOW15-22 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Tang, J, Deng, Z. | | Deposit date: | 2024-06-09 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Multiple independent acquisitions of ACE2 usage in MERS-related coronaviruses.
Cell, 188, 2025
|
|
1UP4
 
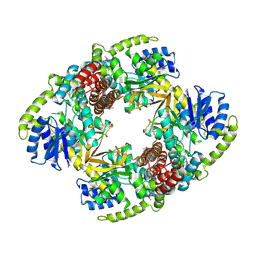 | |
7CGC
 
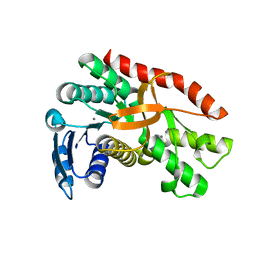 | |
3GZN
 
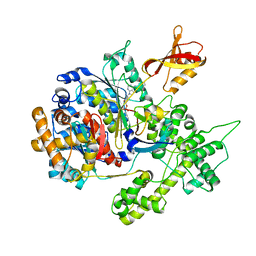 | | Structure of NEDD8-activating enzyme in complex with NEDD8 and MLN4924 | | Descriptor: | NEDD8, NEDD8-activating enzyme E1 catalytic subunit, NEDD8-activating enzyme E1 regulatory subunit, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2009-04-07 | | Release date: | 2010-02-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ.
Mol.Cell, 37, 2010
|
|
3H4N
 
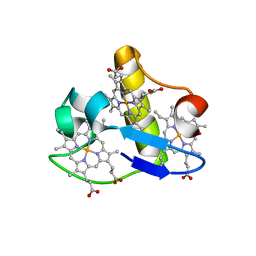 | | PpcD, A cytochrome c7 from Geobacter sulfurreducens | | Descriptor: | Cytochrome c7, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2009-04-20 | | Release date: | 2009-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterization of a family of cytochromes c(7) involved in Fe(III) respiration by Geobacter sulfurreducens.
Biochim.Biophys.Acta, 1797, 2010
|
|
1UP6
 
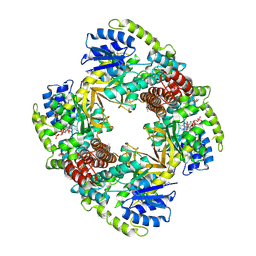 | | Structure of the 6-phospho-beta glucosidase from Thermotoga maritima at 2.55 Angstrom resolution in the tetragonal form with manganese, NAD+ and glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-PHOSPHO-BETA-GLUCOSIDASE, MANGANESE (II) ION, ... | | Authors: | Varrot, A, Yip, V.L, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-09-29 | | Release date: | 2004-08-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | An Unusual Mechanism of Glycoside Hydrolysis Involving Redox and Elimination Steps by a Family 4 Beta-Glycosidase from Thermotoga Maritima.
J.Am.Chem.Soc., 126, 2004
|
|
3H33
 
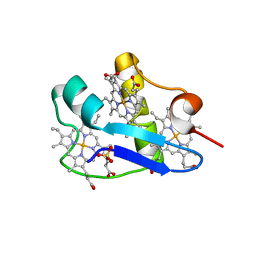 | | PpcC, A cytochrome c7 from Geobacter sulfurreducens | | Descriptor: | Cytochrome c7, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2009-04-15 | | Release date: | 2009-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural characterization of a family of cytochromes c(7) involved in Fe(III) respiration by Geobacter sulfurreducens.
Biochim.Biophys.Acta, 1797, 2010
|
|
3H34
 
 | | PpcE, A cytochrome c7 from Geobacter sulfurreducens | | Descriptor: | Cytochrome c7, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2009-04-15 | | Release date: | 2009-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of a family of cytochromes c(7) involved in Fe(III) respiration by Geobacter sulfurreducens.
Biochim.Biophys.Acta, 1797, 2010
|
|
7FAS
 
 | | VAR2CSA 3D7 ectodomain core region | | Descriptor: | Erythrocyte membrane protein 1, PfEMP1 | | Authors: | Wang, L, Zhaoning, W. | | Deposit date: | 2021-07-07 | | Release date: | 2021-11-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The molecular mechanism of cytoadherence to placental or tumor cells through VAR2CSA from Plasmodium falciparum.
Cell Discov, 7, 2021
|
|
8JKE
 
 | | AfsR(T337A) transcription activation complex | | Descriptor: | DNA(65-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Wang, Y, Zheng, J. | | Deposit date: | 2023-06-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural and functional characterization of AfsR, an SARP family transcriptional activator of antibiotic biosynthesis in Streptomyces.
Plos Biol., 22, 2024
|
|
6DB3
 
 | | JAK3 with Cyanamide CP23 | | Descriptor: | Tyrosine-protein kinase JAK3, [(1S)-1-methyl-6-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-2,3-dihydro-1H-inden-1-yl]cyanamide | | Authors: | Vajdos, F.F. | | Deposit date: | 2018-05-02 | | Release date: | 2018-11-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Identification of Cyanamide-Based Janus Kinase 3 (JAK3) Covalent Inhibitors.
J. Med. Chem., 61, 2018
|
|
6DUD
 
 | | JAK3 with cyanamide CP12 | | Descriptor: | N-[(1S)-6-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-2,3-dihydro-1H-inden-1-yl]imidoformamide, SULFATE ION, Tyrosine-protein kinase JAK3 | | Authors: | Vajdos, F.F. | | Deposit date: | 2018-06-20 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Identification of Cyanamide-Based Janus Kinase 3 (JAK3) Covalent Inhibitors.
J. Med. Chem., 61, 2018
|
|
6DB4
 
 | | JAK3 with Cyanamide CP34 | | Descriptor: | N-[(1S)-6-(5-phenyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)-2,3-dihydro-1H-inden-1-yl]imidoformamide, Tyrosine-protein kinase JAK3 | | Authors: | Vajdos, F.F. | | Deposit date: | 2018-05-02 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Identification of Cyanamide-Based Janus Kinase 3 (JAK3) Covalent Inhibitors.
J. Med. Chem., 61, 2018
|
|
6BJ2
 
 | | TCR589 in complex with HIV(Pol448-456)/HLA-B35 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Jude, K.M, Sibener, L.V, Garcia, K.C. | | Deposit date: | 2017-11-03 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Isolation of a Structural Mechanism for Uncoupling T Cell Receptor Signaling from Peptide-MHC Binding.
Cell, 174, 2018
|
|
6DA4
 
 | | JAK3 with Cyanamide CP10 | | Descriptor: | (Z)-1-{2,2-difluoro-6-[5-(2-methoxyethyl)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]-2,3-dihydro-4H-1,4-benzoxazin-4-yl}methanimine, Tyrosine-protein kinase JAK3 | | Authors: | Vajdos, F.F. | | Deposit date: | 2018-05-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of Cyanamide-Based Janus Kinase 3 (JAK3) Covalent Inhibitors.
J. Med. Chem., 61, 2018
|
|
5U3B
 
 | | Pseudomonas aeruginosa LpxC in complex with NVS-LPXC-01 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-[(2S)-3-amino-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]-4-[(but-2-yn-1-yl)oxy]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Sprague, E.R. | | Deposit date: | 2016-12-01 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Properties of a Potent Inhibitor of Pseudomonas aeruginosa Deacetylase LpxC.
J. Med. Chem., 60, 2017
|
|
5UE8
 
 | | The crystal structure of Munc13-1 C1C2BMUN domain | | Descriptor: | CHLORIDE ION, Protein unc-13 homolog A, ZINC ION | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
5XKN
 
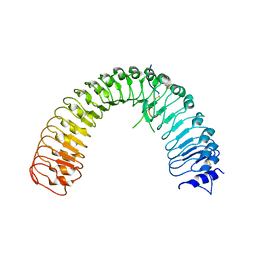 | | Crystal structure of plant receptor ERL2 in complexe with EPFL4 | | Descriptor: | EPIDERMAL PATTERNING FACTOR-like protein 4, LRR receptor-like serine/threonine-protein kinase ERL2 | | Authors: | Chai, J, Lin, G. | | Deposit date: | 2017-05-08 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.651 Å) | | Cite: | A receptor-like protein acts as a specificity switch for the regulation of stomatal development.
Genes Dev., 31, 2017
|
|
7BW6
 
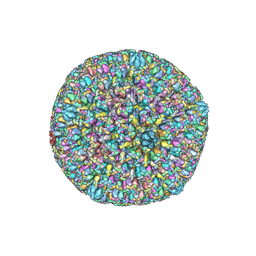 | | Varicella-zoster virus capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Wang, P.Y, Qi, J.X, Liu, C.C, Sun, J.Q. | | Deposit date: | 2020-04-13 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the varicella-zoster virus A-capsid.
Nat Commun, 11, 2020
|
|
2JM5
 
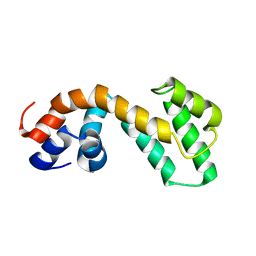 | | Solution Structure of the RGS domain from human RGS18 | | Descriptor: | Regulator of G-protein signaling 18 | | Authors: | Higman, V.A, Leidert, M, Bray, J, Elkins, J, Soundararajan, M, Doyle, D.A, Gileadi, C, Phillips, C, Schoch, G, Yang, X, Brockmann, C, Schmieder, P, Diehl, A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Oschkinat, H, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-11 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2JNU
 
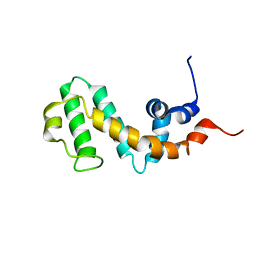 | | Solution structure of the RGS domain of human RGS14 | | Descriptor: | Regulator of G-protein signaling 14 | | Authors: | Dowler, E.F, Diehl, A, Bray, J, Elkins, J, Soundararajan, M, Doyle, D.A, Gileadi, C, Phillips, C, Schoch, G.A, Yang, X, Brockmann, C, Leidert, M, Rehbein, K, Schmieder, P, Kuhne, R, Higman, V.A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Oschkinat, H, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-02 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7C53
 
 | |
