1N5B
 
 | |
8ADE
 
 | |
4KBQ
 
 | | Structure of the CHIP-TPR domain in complex with the Hsc70 Lid-Tail domains | | Descriptor: | E3 ubiquitin-protein ligase CHIP, Heat shock cognate 71 kDa protein | | Authors: | Page, R.C, Amick, J, Nix, J.C, Misra, S. | | Deposit date: | 2013-04-23 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A Bipartite Interaction between Hsp70 and CHIP Regulates Ubiquitination of Chaperoned Client Proteins.
Structure, 23, 2015
|
|
7NIP
 
 | |
4OFQ
 
 | | Structure of the C-terminal domain of the Streptococcus pyogenes antigen I/II-family protein AspA | | Descriptor: | CALCIUM ION, Putative cell surface protein | | Authors: | Hall, M, Nylander, A, Jenkinson, H.F, Persson, K. | | Deposit date: | 2014-01-15 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the C-terminal domain of AspA (antigen I/II-family) protein from Streptococcus pyogenes.
FEBS Open Bio, 4, 2014
|
|
7BJD
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 3 | | Descriptor: | 2-methyl-2-(3-methyl-4-{[4-(methylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino}-1H-pyrazol-1-yl)propanenitrile, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BJH
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 8 | | Descriptor: | CHLORIDE ION, N,N-dimethyl-7H-purin-6-amine, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BJM
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 10 | | Descriptor: | 4-amino-6-{[(2-fluorophenyl)methyl]amino}-7-methyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BJR
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 18 | | Descriptor: | 4-amino-7-methyl-6-[(2R)-2-methylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BJX
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 26 | | Descriptor: | 4-amino-7-methyl-2-[(1-methyl-1H-pyrazol-4-yl)amino]-6-[(2R)-2-methylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BJJ
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 9 | | Descriptor: | 1H-pyrazolo[3,4-d]pyrimidin-4-amine, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BK3
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 45 | | Descriptor: | 4-amino-7-methyl-2-({5-methyl-1-[(3S)-oxolan-3-yl]-1H-pyrazol-4-yl}amino)-6-[(2R)-2-methylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-15 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BJO
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 13 | | Descriptor: | 4-amino-6-(2,3-difluorophenyl)-7-methyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BKO
 
 | | Crystal structure of CHK1 complex with compound 9 | | Descriptor: | 1H-pyrazolo[3,4-d]pyrimidin-4-amine, Serine/threonine-protein kinase Chk1 | | Authors: | Baker, L.M, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-16 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BJE
 
 | | Crystal structure of CHK1-10pt-mutant complex with adenine | | Descriptor: | ADENINE, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BK1
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 32 | | Descriptor: | 4-amino-2-[(1,3-dimethyl-1H-pyrazol-4-yl)amino]-7-methyl-6-[(2R)-2-methylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-15 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BKN
 
 | | Crystal structure of CHK1 complex with adenine | | Descriptor: | ADENINE, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Baker, L.M, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-16 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7N8C
 
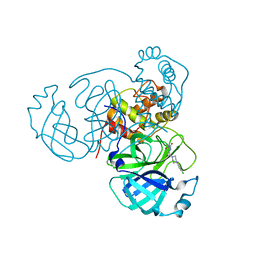 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (Mpro) in complex with Mcule5948770040 | | Descriptor: | 3C-like proteinase, 6-[4-(3,4-dichlorophenyl)piperazin-1-yl]carbonyl-1~{H}-pyrimidine-2,4-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-06-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2.2 Å), X-RAY DIFFRACTION | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
1PBR
 
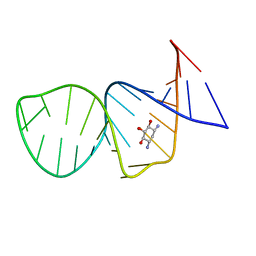 | | STRUCTURE OF 16S RIBOSOMAL RNA, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | 16S RIBOSOMAL RNA, 2,6-diamino-2,6-dideoxy-beta-L-idopyranose-(1-3)-beta-D-ribofuranose, 2-DEOXYSTREPTAMINE, ... | | Authors: | Fourmy, D, Recht, M.I, Blanchard, S, Puglisi, J.D. | | Deposit date: | 1996-09-12 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic.
Science, 274, 1996
|
|
7BK2
 
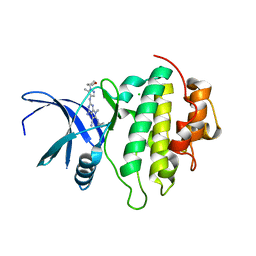 | | Crystal structure of CHK1-10pt-mutant complex with compound 44 | | Descriptor: | 4-amino-7-methyl-2-({5-methyl-1-[(3R)-oxolan-3-yl]-1H-pyrazol-4-yl}amino)-6-[(2R)-2-methylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-15 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
8P1O
 
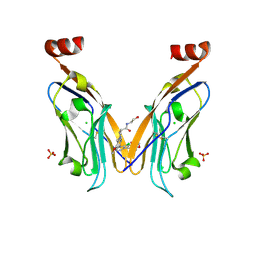 | | Solubilizer tag effect on PD-L1/inhibitor binding properties for m-terphenyl derivatives | | Descriptor: | (3~{R})-1-[[4-[2-chloranyl-3-(2,3-dihydro-1,4-benzodioxin-6-yl)phenyl]-2-methoxy-phenyl]methyl]-~{N}-(2-hydroxyethyl)pyrrolidine-3-carboxamide, CHLORIDE ION, Programmed cell death 1 ligand 1, ... | | Authors: | Plewka, J, Magiera-Mularz, K, Surmiak, E, Kalinowska-Tluscik, J, Holak, T.A. | | Deposit date: | 2023-05-12 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Solubilizer Tag Effect on PD-L1/Inhibitor Binding Properties for m -Terphenyl Derivatives.
Acs Med.Chem.Lett., 15, 2024
|
|
8BJH
 
 | | chimera of the inactive ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, with the double mutation K3528M and K3535I, fused to a proline-Rich-Domain (PRD) and profilin, bound to Latrunculin B-ADP-Mg-actin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
8BO1
 
 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, bound to Latrunculin-B-ATP-Mg-actin, and 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE and 2 Mg ions | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AZIDE ION, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
8BJI
 
 | | chimera of ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo fused to a proline-Rich-Domain (PRD) and profilin, bound to ADP-Mg-actin and a sulfate ion | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
8BR1
 
 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, bound to Latrunculin-B-ATP-Mg-actin, and 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE and 2 Mg ions | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-22 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.044 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
