2XP5
 
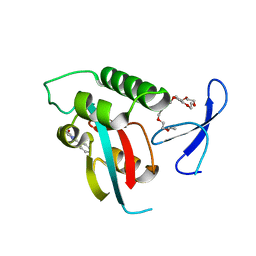 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 5-METHYL-2-PHENYL-1H-IMIDAZOLE-4-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XYS
 
 | | Crystal structure of Aplysia californica AChBP in complex with strychnine | | Descriptor: | SOLUBLE ACETYLCHOLINE RECEPTOR, STRYCHNINE | | Authors: | Brams, M, Pandya, A, Kuzmin, D, van Elk, R, Krijnen, L, Yakel, J.L, Tsetlin, V, Smit, A.B, Ulens, C. | | Deposit date: | 2010-11-19 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | A Structural and Mutagenic Blueprint for Molecular Recognition of Strychnine and D-Tubocurarine by Different Cys-Loop Receptors.
Plos Biol., 9, 2011
|
|
2XYQ
 
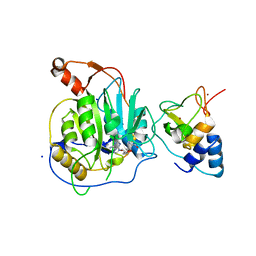 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
2XZG
 
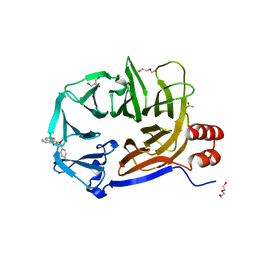 | | Clathrin Terminal Domain Complexed with Pitstop 1 | | Descriptor: | 2-(4-AMINOBENZYL)-1,3-DIOXO-2,3-DIHYDRO-1H-BENZO[DE]ISOQUINOLINE-5-SULFONATE, ACETATE ION, CLATHRIN HEAVY CHAIN 1, ... | | Authors: | Bulut, H, Von Kleist, L, Saenger, W, Haucke, V. | | Deposit date: | 2010-11-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of the Clathrin Terminal Domain in Regulating Coated Pit Dynamics Revealed by Small Molecule Inhibition.
Cell(Cambridge,Mass.), 146, 2011
|
|
2ZAV
 
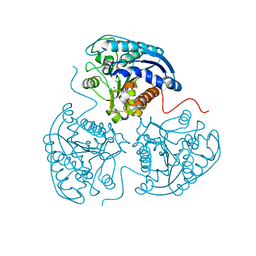 | |
2ZCI
 
 | |
2XPA
 
 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 4-[(2-amino-2-oxoethyl)(methyl)carbamoyl]-2-phenyl-1H-imidazole-5-carboxylic acid, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XYT
 
 | | Crystal structure of Aplysia californica AChBP in complex with d- tubocurarine | | Descriptor: | D-TUBOCURARINE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Brams, M, Pandya, A, Kuzmin, D, van Elk, R, Krijnen, L, Yakel, J.L, Tsetlin, V, Smit, A.B, Ulens, C. | | Deposit date: | 2010-11-19 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Structural and Mutagenic Blueprint for Molecular Recognition of Strychnine and D-Tubocurarine by Different Cys-Loop Receptors.
Plos Biol., 9, 2011
|
|
2XS6
 
 | | CRYSTAL STRUCTURE OF THE RHOGAP DOMAIN OF HUMAN PIK3R2 | | Descriptor: | CHLORIDE ION, PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT BETA | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, van der Berg, S, Wahlberg, E, Weigelt, J, Nordlund, P. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Structure and Catalytic Mechanism of Human Sphingomyelin Phosphodiesterase Like 3A - an Acid Sphingomyelinase Homolog with a Novel Nucleotide Hydrolase Activity.
FEBS J., 283, 2016
|
|
2XRQ
 
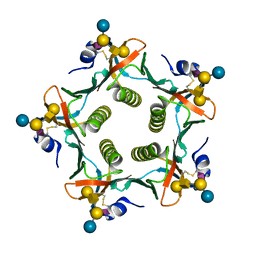 | | Crystal structures exploring the origins of the broader specificity of escherichia coli heat-labile enterotoxin compared to cholera toxin | | Descriptor: | HEAT-LABILE ENTEROTOXIN B CHAIN, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Holmner, A, Mackenzie, A, Okvist, M, Jansson, L, Lebens, M, Teneberg, S, Krengel, U. | | Deposit date: | 2010-09-19 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures Exploring the Origins of the Broader Specificity of Escherichia Coli Heat-Labile Enterotoxin Compared to Cholera Toxin
J.Mol.Biol., 406, 2011
|
|
3CAS
 
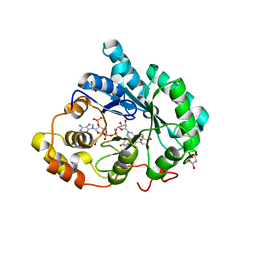 | | Crystal structure of 5beta-reductase (AKR1D1) in complex with NADP+ and 4-androstenedione | | Descriptor: | 1,2-ETHANEDIOL, 3-oxo-5-beta-steroid 4-dehydrogenase, 4-ANDROSTENE-3-17-DIONE, ... | | Authors: | Faucher, F, Cantin, L, Breton, R. | | Deposit date: | 2008-02-20 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Human Delta4-3-Ketosteroid 5beta-Reductase (AKR1D1) Reveal the Presence of an Alternative Binding Site Responsible for Substrate Inhibition (dagger) (,) (double dagger).
Biochemistry, 47, 2008
|
|
2XO2
 
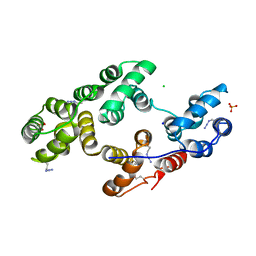 | | Human Annexin V with incorporated Methionine analogue Azidohomoalanine | | Descriptor: | ANNEXIN A5, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Debela, M, Merkel, L, Goettig, P, Budisa, N. | | Deposit date: | 2010-08-09 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Non-Canonical Amino Acids for Click Chemistry Reactions Incorporated in Human Annexin V
To be Published
|
|
3CBQ
 
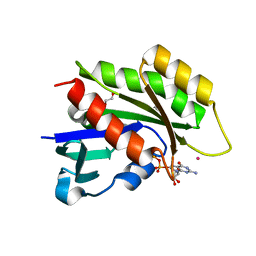 | | Crystal structure of the human REM2 GTPase with bound GDP | | Descriptor: | 1,2-ETHANEDIOL, GTP-binding protein REM 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nedyalkova, L, Shen, Y, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-22 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of the human REM2 GTPase with bound GDP.
To be Published
|
|
2XNI
 
 | | Protein-ligand complex of a novel macrocyclic HCV NS3 protease inhibitor derived from amino cyclic boronates | | Descriptor: | (1-{[(10-tert-butyl-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19,23,23a-tetradecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosin-7(3H)-yl)carbonyl]amino}-3-hydroxypropyl)(trihydroxy)borate(1-), MAGNESIUM ION, NS3 PROTEASE, ... | | Authors: | Li, X, Zhang, Y.-K, Liu, Y, Ding, C.Z, Zhou, Y, Li, Q, Plattner, J.J, Baker, S.J, Zhang, S, Kazmierski, W.M, Wright, L.L, Smith, G.K, Grimes, R.M, Crosby, R.M, Creech, K.L, Carballo, L.H, Slater, M.J, Jarvest, R.L, Thommes, P, Hubbard, J.A, Convery, M.A, Nassau, P.M, McDowell, W, Skarzynski, T.J, Qian, X, Fan, D, Liao, L, Ni, Z.-J, Pennicott, L.E, Zou, W, Wright, J. | | Deposit date: | 2010-08-02 | | Release date: | 2011-08-17 | | Last modified: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Novel Macrocyclic Hcv Ns3 Protease Inhibitors Derived from Alpha-Amino Cyclic Boronates.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XUA
 
 | | Crystal structure of the enol-lactonase from Burkholderia xenovorans LB400 | | Descriptor: | 3-OXOADIPATE ENOL-LACTONASE, LAEVULINIC ACID | | Authors: | Bains, J, Kaufman, L, Farnell, B, Boulanger, M.J. | | Deposit date: | 2010-10-17 | | Release date: | 2011-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Product Analog Bound Form of 3-Oxoadipate-Enol- Lactonase (Pcad) Reveals a Multifunctional Role for the Divergent CAP Domain.
J.Mol.Biol., 406, 2011
|
|
2XVT
 
 | | Structure of the extracellular domain of human RAMP2 | | Descriptor: | CALCIUM ION, RECEPTOR ACTIVITY-MODIFYING PROTEIN 2 | | Authors: | Quigley, A, Pike, A.C.W, Burgess-Brown, N, Krojer, T, Shrestha, L, Goubin, S, Kim, J, Das, S, Muniz, J.R.C, Canning, P, Chaikuad, A, Vollmar, M, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bountra, C, Barr, A.J, Carpenter, E.P. | | Deposit date: | 2010-10-31 | | Release date: | 2010-12-29 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the Extracellular Domain of Human Ramp2
To be Published
|
|
3CCD
 
 | | 1.0 A Structure of Post-Succinimide His15Asp HPr | | Descriptor: | Phosphocarrier protein HPr, SULFATE ION | | Authors: | Napper, S, Prasad, L, Delbaere, L.T.J. | | Deposit date: | 2008-02-25 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural investigation of a phosphorylation-catalyzed, isoaspartate-free, protein succinimide: crystallographic structure of post-succinimide His15Asp histidine-containing protein
Biochemistry, 47, 2008
|
|
2XPB
 
 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 5-[BENZYL(METHYL)CARBAMOYL]-2-(3-CHLOROPHENYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XQZ
 
 | | Neutron structure of the perdeuterated Toho-1 R274N R276N double mutant beta-lactamase | | Descriptor: | BETA-LACTAMSE TOHO-1 | | Authors: | Tomanicek, S.J, Wang, K.K, Weiss, K.L, Blakeley, M.P, Cooper, J, Chen, Y, Coates, L. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-22 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | The Active Site Protonation States of Perdeuterated Toho-1 Beta-Lactamase Determined by Neutron Diffraction Support a Role for Glu166 as the General Base in Acylation.
FEBS Lett., 585, 2011
|
|
2XUR
 
 | | The G157C mutation in the Escherichia coli sliding clamp specifically affects initiation of replication | | Descriptor: | DNA POLYMERASE III SUBUNIT BETA | | Authors: | Johnsen, L, Morigen, Dalhus, B, Bjoras, M, Flaatten, I, Waldminghaus, T, Skarstad, K. | | Deposit date: | 2010-10-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The G157C Mutation in the Escherichia Coli Sliding Clamp Specifically Affects Initiation of Replication.
Mol.Microbiol., 79, 2011
|
|
3CE2
 
 | | Crystal structure of putative peptidase from Chlamydophila abortus | | Descriptor: | Putative peptidase, ZINC ION | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Eberle, M, Maletic, M, Meyer, A.J, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-28 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of putative peptidase from Chlamydophila abortus.
To be Published
|
|
3C8K
 
 | | The crystal structure of Ly49C bound to H-2Kb | | Descriptor: | H-2 class I histocompatibility antigen, K-B alpha chain, Natural killer cell receptor Ly-49C, ... | | Authors: | Deng, L, Mariuzza, R.A. | | Deposit date: | 2008-02-12 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular architecture of the major histocompatibility complex class I-binding site of Ly49 natural killer cell receptors.
J.Biol.Chem., 283, 2008
|
|
3CDK
 
 | | Crystal structure of the co-expressed succinyl-CoA transferase A and B complex from Bacillus subtilis | | Descriptor: | Succinyl-CoA:3-ketoacid-coenzyme A transferase subunit A, Succinyl-CoA:3-ketoacid-coenzyme A transferase subunit B | | Authors: | Kim, Y, Zhou, M, Stols, L, Eschenfeldt, W, Donnelly, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-27 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of the co-expressed succinyl-CoA transferase A and B complex from Bacillus subtilis.
To be Published
|
|
2XRS
 
 | | Crystal structures exploring the origins of the broader specificity of escherichia coli heat-labile enterotoxin compared to cholera toxin | | Descriptor: | HEAT-LABILE ENTEROTOXIN B CHAIN, beta-D-galactopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Holmner, A, Mackenzie, A, Okvist, M, Jansson, L, Lebens, M, Teneberg, S, Krengel, U. | | Deposit date: | 2010-09-20 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structures Exploring the Origins of the Broader Specificity of Escherichia Coli Heat-Labile Enterotoxin Compared to Cholera Toxin
J.Mol.Biol., 406, 2011
|
|
2XX9
 
 | | Crystal structure of 1-((2-fluoro-4-(3-(trifluoromethyl)-4,5,6,7- tetrahydro-1H-indazol-1-yl)phenyl)methyl)-2-pyrrolidinone in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N,N-DIMETHYL-4-[3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOL-1-YL]BENZAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-06 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
