4YLC
 
 | |
3WHL
 
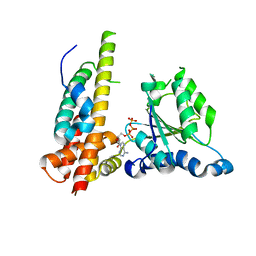 | | Crystal structure of Nas2 N-terminal domain complexed with PAN-Rpt5C chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Probable 26S proteasome regulatory subunit p27, Proteasome-activating nucleotidase, ... | | Authors: | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | Deposit date: | 2013-08-26 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3WHJ
 
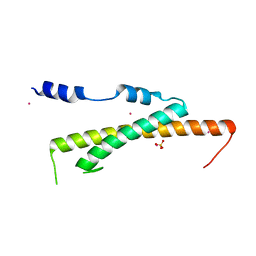 | | Crystal structure of Nas2 N-terminal domain | | Descriptor: | CADMIUM ION, Probable 26S proteasome regulatory subunit p27, SULFATE ION | | Authors: | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | Deposit date: | 2013-08-26 | | Release date: | 2014-03-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3WHK
 
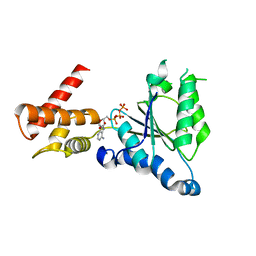 | | Crystal structure of PAN-Rpt5C chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Proteasome-activating nucleotidase, 26S protease regulatory subunit 6A | | Authors: | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | Deposit date: | 2013-08-26 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
8GZB
 
 | | SARS-CoV-2 3CLpro | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-chlorophenyl)-1,3,4-oxadiazole, 3C-like proteinase nsp5 | | Authors: | Wang, F, Cen, Y.X, Tian, P. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nature-inspired catalytic asymmetric rearrangement of cyclopropylcarbinyl cation.
Sci Adv, 9, 2023
|
|
8KE3
 
 | | PylRS C-terminus domain mutant bound with D-3-trifluoromethylphenylalanine and AMPNP | | Descriptor: | (2R)-2-azanyl-3-[3-(trifluoromethyl)phenyl]propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89980984 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE2
 
 | | PylRS C-terminus domain mutant bound with L-3-trifluoromethylphenylalanine and AMPNP | | Descriptor: | (2S)-2-azanyl-3-[3-(trifluoromethyl)phenyl]propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.19639969 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE5
 
 | | PylRS C-terminus domain mutant bound with D-3-chlorophenylalanine and AMPNP | | Descriptor: | (2R)-2-azanyl-3-(3-chlorophenyl)propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.900073 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE1
 
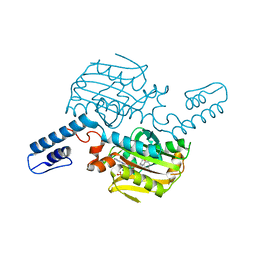 | | PylRS C-terminus domain mutant bound with L-3-bromophenylalanine and AMPNP | | Descriptor: | 3-bromo-L-phenylalanine, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.50081539 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE6
 
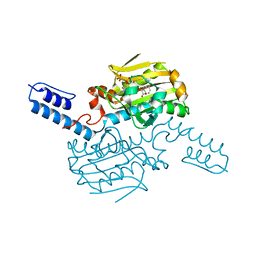 | | PylRS C-terminus domain mutant bound with L-3-chlorophenylalanine and AMPNP | | Descriptor: | 3-CHLORO-L-PHENYLALANINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89570856 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE4
 
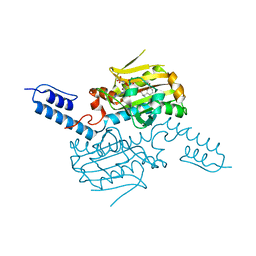 | | PylRS C-terminus domain mutant bound with D-3-bromophenylalanine and AMPNP | | Descriptor: | (2R)-2-azanyl-3-(3-bromophenyl)propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75050962 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
4ZGY
 
 | |
2MVW
 
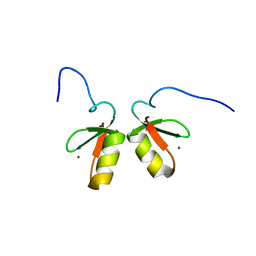 | | Solution structure of the TRIM19 B-box1 (B1) of human promyelocytic leukemia (PML) | | Descriptor: | Protein PML, ZINC ION | | Authors: | Huang, S, Naik, M.T, Fan, P, Wang, Y, Chang, C, Huang, T. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The B-box 1 dimer of human promyelocytic leukemia protein.
J.Biomol.Nmr, 60, 2014
|
|
4ZGZ
 
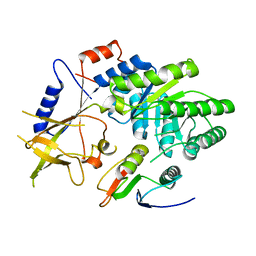 | |
3OSM
 
 | |
3OSE
 
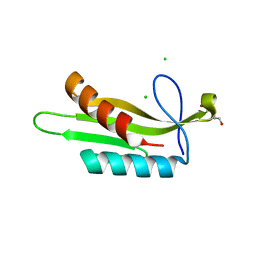 | |
3OST
 
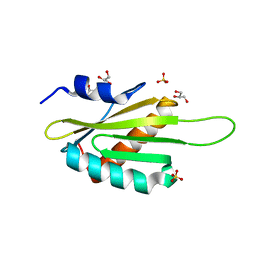 | | Structure of the Kinase Associated-1 (KA1) from Kcc4p | | Descriptor: | GLYCEROL, SULFATE ION, serine/threonine-protein kinase KCC4 | | Authors: | Moravcevic, K, Lemmon, M.A. | | Deposit date: | 2010-09-09 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Kinase Associated-1 Domains Drive MARK/PAR1 Kinases to Membrane Targets by Binding Acidic Phospholipids.
Cell(Cambridge,Mass.), 143, 2010
|
|
7VIJ
 
 | | Crystal structure of USP7-HUBL domain | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Feng, N, Zeng, K.W. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Neuroinflammation inhibition by small-molecule targeting USP7 noncatalytic domain for neurodegenerative disease therapy.
Sci Adv, 8, 2022
|
|
7XPY
 
 | | Crystal structure of USP7 in complex with its inhibitor | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, [(3S,3aR,4R,6Z,9S,10E,11aR)-9-acetyloxy-6-(acetyloxymethyl)-3,10-dimethyl-2-oxidanylidene-3a,4,5,8,9,11a-hexahydro-3H-cyclodeca[b]furan-4-yl] (E)-2-methyl-4-oxidanyl-but-2-enoate | | Authors: | Feng, N, Zeng, K.W. | | Deposit date: | 2022-05-06 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Neuroinflammation inhibition by small-molecule targeting USP7 noncatalytic domain for neurodegenerative disease therapy.
Sci Adv, 8, 2022
|
|
7E0N
 
 | | Crystal structure of Monoacylglycerol Lipase chimera | | Descriptor: | GLYCEROL, Thermostable monoacylglycerol lipase,Lipase | | Authors: | Lan, D.M. | | Deposit date: | 2021-01-28 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Monoacylglycerol Lipase chimera
To Be Published
|
|
